Measurement of $CP$ asymmetry in $B^0_s \rightarrow D^{\mp}_s K^{\pm}$ decays
[to restricted-access page]Abstract
A measurement of the $ C P$ -violating parameters in $ B^0_s \rightarrow D_s^\mp K^\pm$ decays obtained from $pp$ collisions is reported, using a data set corresponding to an integrated luminosity of $6 \text{ fb} ^{-1} $ at a centre-of-mass energy of $\sqrt{s} = 13 \text{ Te V} $ recorded with the LHCb detector between 2015 and 2018. The measured values are: \begin{align*} C_f &= \phantom{+}0.791 \pm 0.061 \pm 0.022 , \\ A^{\Delta \Gamma}_f &= -0.051 \pm 0.134 \pm 0.037 ,\\ A^{\Delta \Gamma}_{\bar{f}} &= -0.303 \pm 0.125 \pm 0.036 ,\\ S_f &= -0.571 \pm 0.084 \pm 0.023 ,\\ S_{\bar{f}} &= -0.503 \pm 0.084 \pm 0.025 , \end{align*} where the uncertainties are statistical and systematic, respectively. Together with the value of the $ B ^0_ s $ mixing phase $-2\beta_{ s } $, these parameters are used to obtain a measurement of the CKM angle $\gamma$ from $ B^0_s \rightarrow D_s^\mp K^\pm$ decays of $\gamma = ( 74 \pm 11)^{\circ}$ modulo $180^{\circ}$, where the uncertainty contains both statistical and systematic contributions.
Figures and captions
|
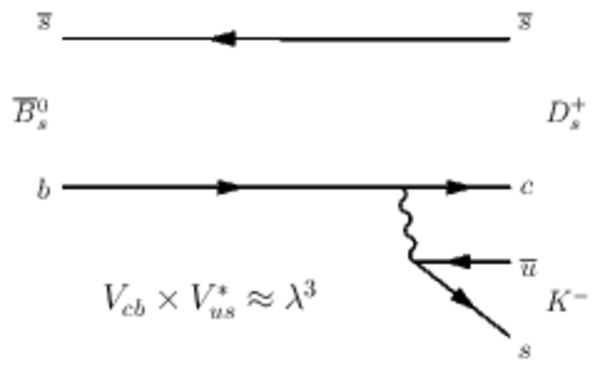
Feynman diagrams for $ B ^0_ s \rightarrow D_s^+ K^-$ decays (left) without and (right) with $ B ^0_ s $ -- $\overline{ B } {}^0_ s $ mixing. |
Fig1a.pdf [57 KiB] HiDef png [26 KiB] Thumbnail [15 KiB] *.C file |
|
|
Fig1b.pdf [59 KiB] HiDef png [42 KiB] Thumbnail [25 KiB] *.C file |

|
|
|
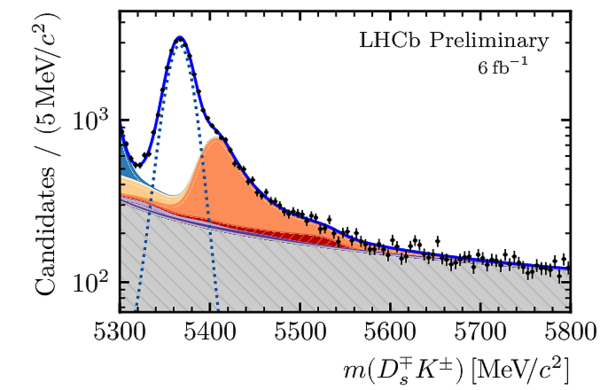
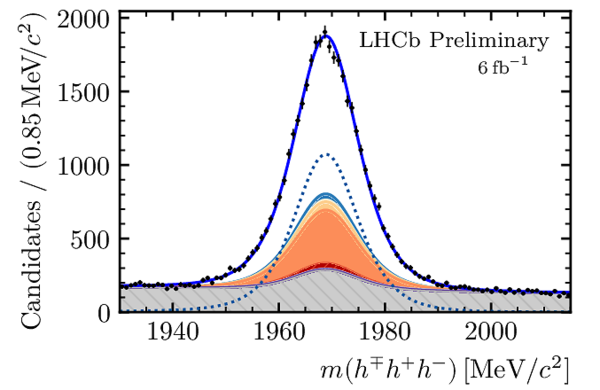
Distributions of the (left) $ D ^{\pm}_ s K^{\mp}$ and (right) $3h$ invariant masses for selected $D_s^\mp K^\pm$ candidates. In each plot, the contributions from all $ D ^-_ s $ final states and years of data taking are combined. The solid blue curve is the total result of the simultaneous fit. The dotted curve shows the $ B^0_s \rightarrow D_s^\mp K^\pm$ signal and the shaded stacked histograms show the different background contributions. |
legend[..].pdf [11 KiB] HiDef png [53 KiB] Thumbnail [44 KiB] *.C file |

|
|
mdfit_[..].pdf [154 KiB] HiDef png [294 KiB] Thumbnail [193 KiB] *.C file |

|
|
|
mdfit_[..].pdf [154 KiB] HiDef png [341 KiB] Thumbnail [216 KiB] *.C file |
|
|
|
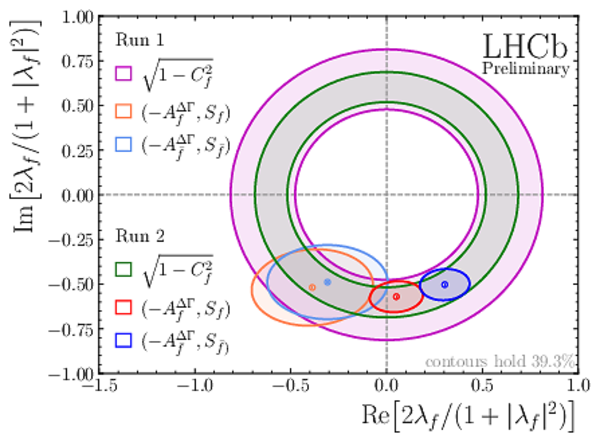
Representation of the $ C P$ -violating parameters in the $\mathrm{Im} \left[2\lambda_f/(1+|\lambda_f|^2)\right]$ vs $\mathrm{Re} \left[2\lambda_f/(1+|\lambda_f|^2)\right]$ cartesian plane. |
dsk_meas.pdf [246 KiB] HiDef png [542 KiB] Thumbnail [383 KiB] *.C file |
|
|
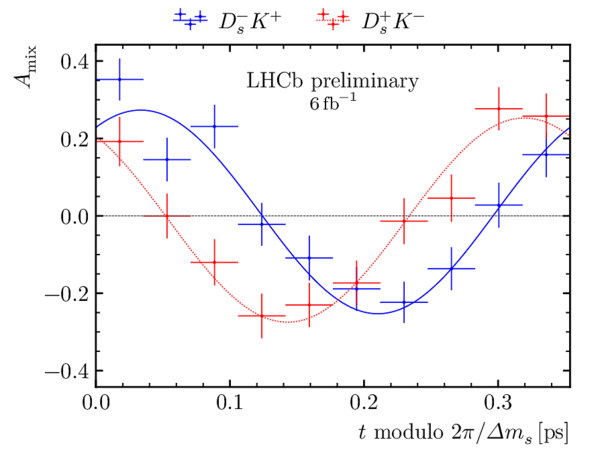
(Top) Decay-time distribution of $ B^0_s \rightarrow D_s^\mp K^\pm$ candidates obtained by the sPlot technique. (Bottom) $ C P$ -asymmetry for the (blue) $ D ^-_ s K^+$ and the (red) $ D ^+_ s K^-$ final states, folded into one mixing period, $2\pi/\Delta m_{ s } $. In both plots, the solid curve shows the result of the decay-time fit. |
oscill[..].pdf [21 KiB] HiDef png [131 KiB] Thumbnail [106 KiB] *.C file |

|
|
asymme[..].pdf [20 KiB] HiDef png [168 KiB] Thumbnail [121 KiB] *.C file |
|
|
|
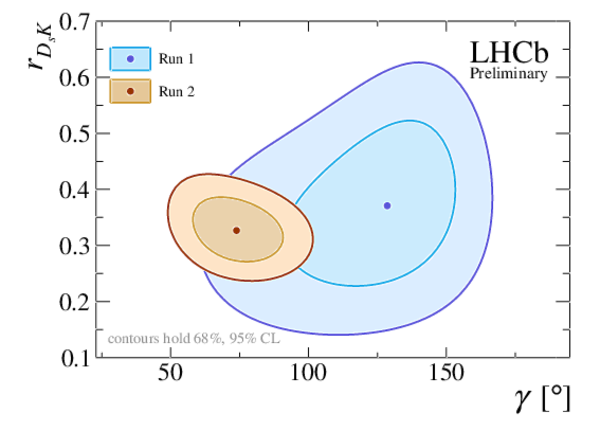
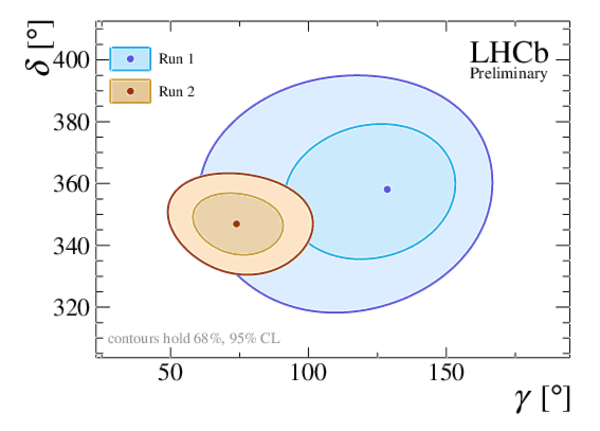
Profile likelihood contours of (top left) $ r_{D_sK}$ vs. $\gamma $ , and (top right) $\delta$ vs. $\gamma $ . The markers denote the best-fit values. The contours correspond to 68.3% CL (95.5% CL). The bottom plot shows $ 1-{\rm CL}$ for the angle $\gamma $ , together with the central value and the 68.3% CL interval as obtained from the frequentist method described in the text. The results with the Run 2 and Run 1 labels are from the present analysis and from Ref. [20], respectively. |
gammac[..].pdf [17 KiB] HiDef png [324 KiB] Thumbnail [189 KiB] *.C file |
|
|
gammac[..].pdf [17 KiB] HiDef png [317 KiB] Thumbnail [188 KiB] *.C file |
|
|
|
gammac[..].pdf [15 KiB] HiDef png [274 KiB] Thumbnail [183 KiB] *.C file |

|
|
|
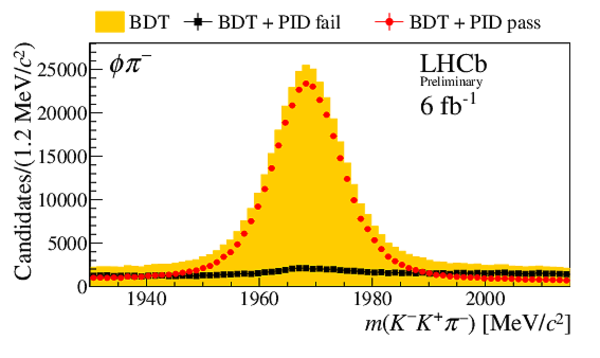
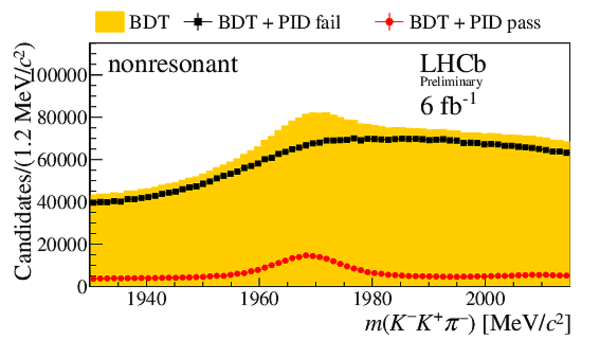
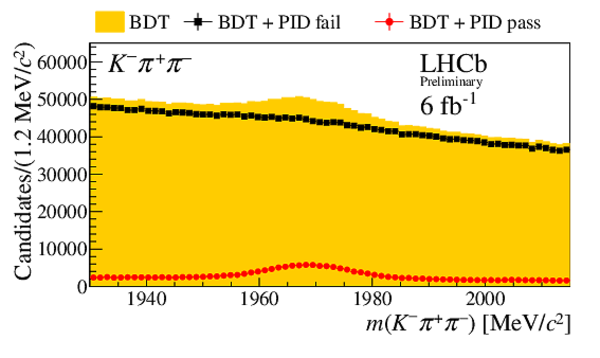
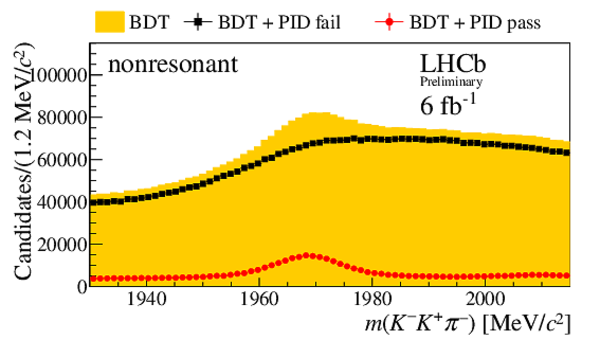
Invariant mass distribution $m(h^{-}h^{+}h^{\pm})$ after BDT selection (orange) and for candidates passing (red circles) or failing (black squares) the PID selection on the $ D ^-_ s $ candidate decay products. No PID requirements are applied to the companion kaon candidate. Distributions are shown for the final states (top left) $ D ^-_ s \rightarrow \phi\pi ^- $ , (top right) $ D ^-_ s \rightarrow K^{*0} K ^- $ , (middle left) $ D_s^- \rightarrow ( K ^- K ^+ \pi ^- )_{\text{nonresonant}}$, (middle right) $ D ^-_ s \rightarrow K ^- \pi ^+ \pi ^- $ , and (bottom) $ D ^-_ s \rightarrow \pi ^- \pi ^+ \pi ^- $ . These subsamples are defined by the invariant mass selections on the $K K$ and $K \pi$ systems given in Sec. 3. |
pid_ch[..].pdf [19 KiB] HiDef png [190 KiB] Thumbnail [160 KiB] *.C file |

|
|
pid_ch[..].pdf [20 KiB] HiDef png [214 KiB] Thumbnail [184 KiB] *.C file |

|
|
|
pid_ch[..].pdf [19 KiB] HiDef png [200 KiB] Thumbnail [170 KiB] *.C file |
|
|
|
pid_ch[..].pdf [19 KiB] HiDef png [201 KiB] Thumbnail [170 KiB] *.C file |

|
|
|
pid_ch[..].pdf [19 KiB] HiDef png [220 KiB] Thumbnail [190 KiB] *.C file |

|
|
|
The decay-time distribution of $ B^0_s \rightarrow D_s^\mp K^\pm$ signal candidates obtained using the sPlot technique. The dotted blue curve (left axis, arbitrary units) describes the decay rate. The solid orange curve (right axis, arbitrary units) describes the decay-time acceptance by which the decay rate is multiplied. |
accept[..].pdf [14 KiB] HiDef png [183 KiB] Thumbnail [124 KiB] *.C file |

|
|
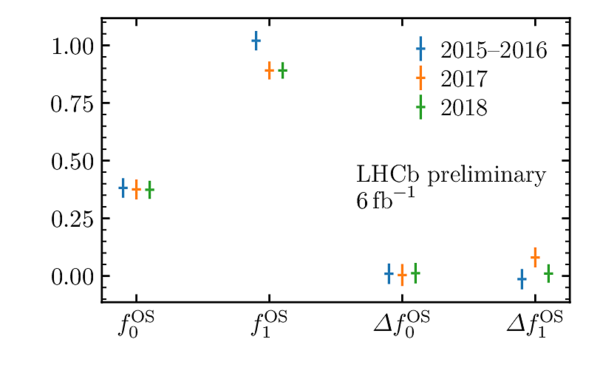
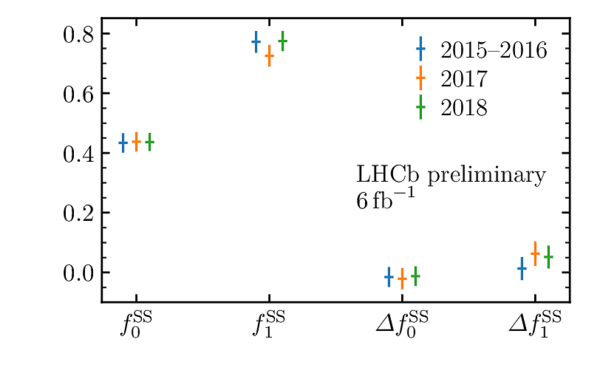
Flavour tagging calibration parameters for (left) the OS combination and (right) the SS combination obtained from the decay time fit to $ B^0_s \rightarrow D_s^- \pi^+$ . Uncertainties shown are statistical only, and are scaled by a factor of 10 for $f_{0}$ and $\Delta f_{0}$. These calibrations use a similar method to that described in Ref. [40], but use an updated selection and resolution determination for the analysis described here. |
OS_cal[..].pdf [12 KiB] HiDef png [107 KiB] Thumbnail [89 KiB] *.C file |
|
|
SS_cal[..].pdf [11 KiB] HiDef png [100 KiB] Thumbnail [82 KiB] *.C file |
|
|
|
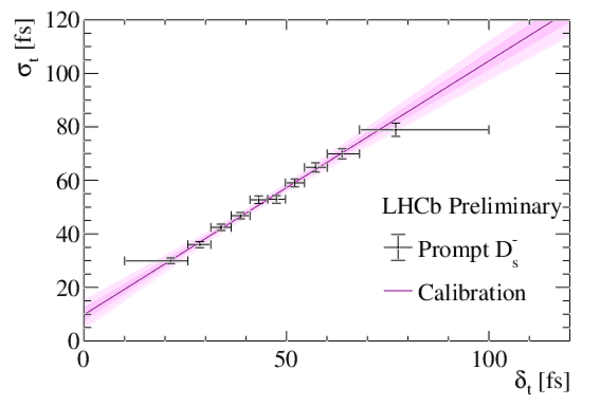
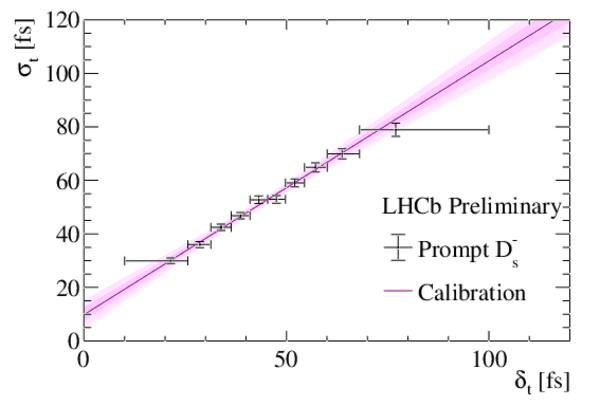
Measured decay-time resolution $\sigma_t$ as a function of the per-candidate decay-time uncertainty $\delta_t$ for prompt $D_s^{\mp}$ candidates combined with a random track. Data points are from the data collected in 2018 only. The solid line shows the linear fit to the data, as discussed in the text. The confidence intervals corresponding to one and two standard deviations of statistical uncertainty are indicated in magenta. |
Prompt[..].pdf [450 KiB] HiDef png [276 KiB] Thumbnail [162 KiB] *.C file |
|
|
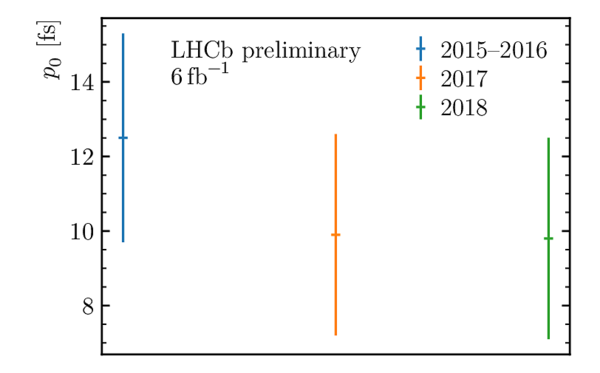
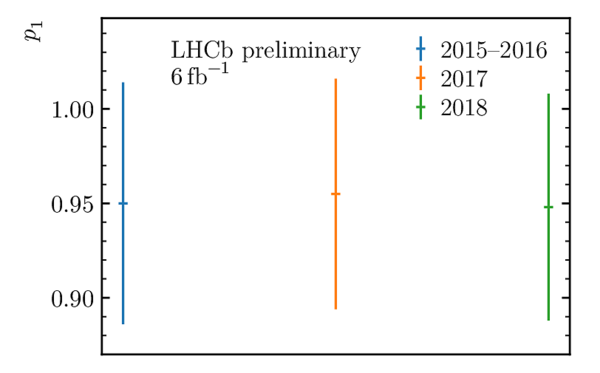
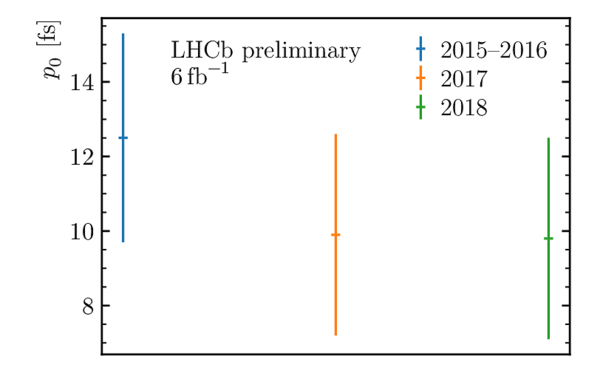
Decay-time resolution calibration parameters $p_0$ (left) and $p_1$ (right) used in the decay-time fit, obtained by combining the two Gaussian widths forming the core of the prompt $ D ^-_ s $ $\pi ^+$ decay-time distribution. |
resolu[..].pdf [10 KiB] HiDef png [72 KiB] Thumbnail [57 KiB] *.C file |
|
|
resolu[..].pdf [9 KiB] HiDef png [75 KiB] Thumbnail [57 KiB] *.C file |

|
|
|
Distributions of the (left) $ D ^{\pm}_ s K^{\mp}$ and (right) $3h$ invariant masses for $ B^0_s \rightarrow D_s^\mp K^\pm$ final states, presented using a logarithmic scale on the vertical axis. In each plot, the contributions from all $ D ^-_ s $ final states and years of data taking are combined. The solid blue curve is the total result of the simultaneous fit. The dotted curve shows the $ B^0_s \rightarrow D_s^\mp K^\pm$ signal and the fully coloured stacked histograms show the different background contributions. |
legend[..].pdf [11 KiB] HiDef png [53 KiB] Thumbnail [44 KiB] *.C file |

|
|
mdfit_[..].pdf [163 KiB] HiDef png [408 KiB] Thumbnail [267 KiB] *.C file |

|
|
|
mdfit_[..].pdf [163 KiB] HiDef png [448 KiB] Thumbnail [285 KiB] *.C file |

|
|
|
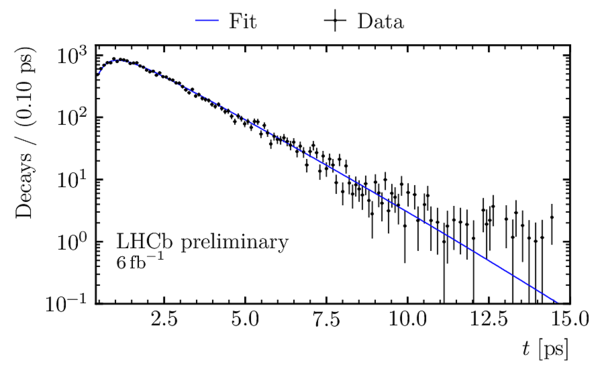
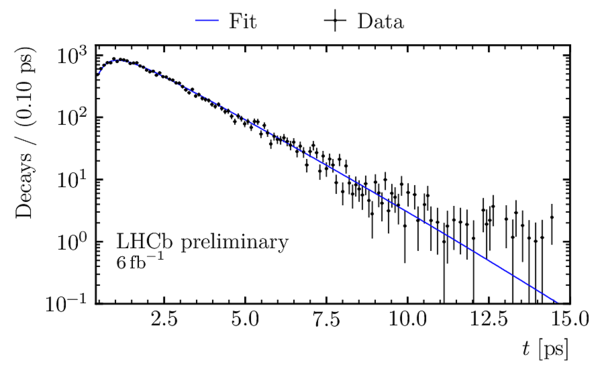
Decay-time distribution of $ B^0_s \rightarrow D_s^\mp K^\pm$ candidates obtained using the sPlot technique, with a logarithmic scale on the $y$ axis. The decay-time fit is overlaid. |
oscill[..].pdf [22 KiB] HiDef png [148 KiB] Thumbnail [126 KiB] *.C file |
|
|
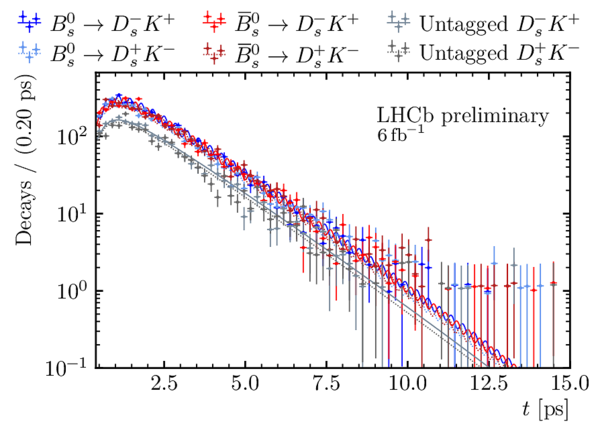
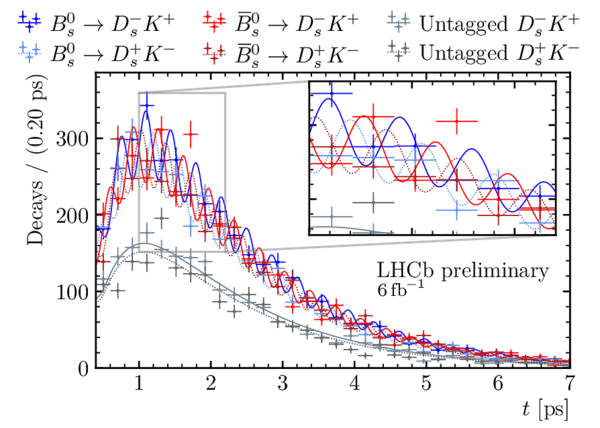
Decay-time distribution of signal $ B^0_s \rightarrow D_s^\mp K^\pm$ candidates split by final state and tagging category. |
oscill[..].pdf [81 KiB] HiDef png [556 KiB] Thumbnail [320 KiB] *.C file |
|
|
oscill[..].pdf [75 KiB] HiDef png [431 KiB] Thumbnail [246 KiB] *.C file |

|
|
|
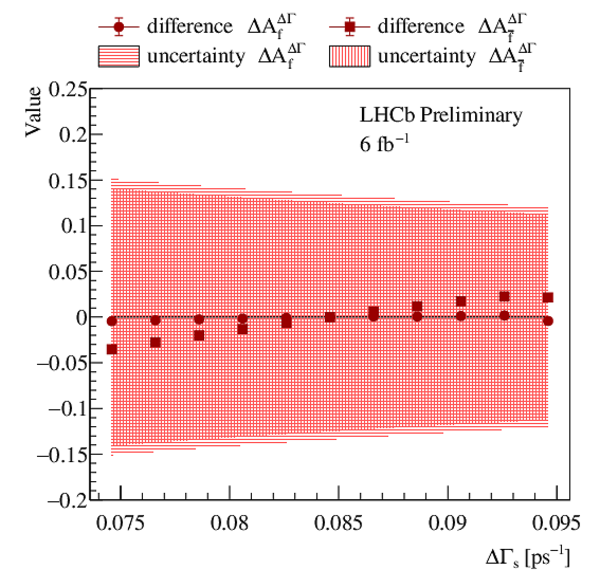
Dependence of the parameters $ A^{\Delta \Gamma}_f$ and $ A^{\Delta \Gamma}_{\bar{f}}$ on $\Delta\Gamma_{ s }$ . Red horizontally and vertically hatched bands represent the statistical uncertainty of the $ A^{\Delta \Gamma}_f$ and $ A^{\Delta \Gamma}_{\bar{f}}$ parameters, while circles (squares) denote the difference of the $ A^{\Delta \Gamma}_f$ ( $ A^{\Delta \Gamma}_{\bar{f}}$ ) parameter with respect to the baseline result. |
scan_d[..].pdf [14 KiB] HiDef png [709 KiB] Thumbnail [412 KiB] *.C file |
|
|
Animated gif made out of all figures. |
CONF-2023-004.gif Thumbnail |

|
Tables and captions
|
Values of the $ C P$ observables obtained from the fit to the decay-time distribution of $ B^0_s \rightarrow D_s^\mp K^\pm$ decays. The first uncertainty is statistical and the second is systematic. |
Table_1.pdf [59 KiB] HiDef png [91 KiB] Thumbnail [42 KiB] tex code |

|
|
Statistical correlation matrix of the $ C P$ observables. Other fit parameters have negligible correlations with the $ C P$ observables. |
Table_2.pdf [58 KiB] HiDef png [40 KiB] Thumbnail [19 KiB] tex code |

|
|
Systematic uncertainties on the $ C P$ observables, relative to the statistical uncertainties. |
Table_3.pdf [57 KiB] HiDef png [111 KiB] Thumbnail [47 KiB] tex code |

|
|
Correlation matrix of the total systematic uncertainties of the $ C P$ observables. |
Table_4.pdf [58 KiB] HiDef png [40 KiB] Thumbnail [19 KiB] tex code |

|
Supplementary Material [file]
Created on 26 April 2024.