Information
LHCb-PAPER-2014-028
CERN-PH-EP-2014-182
arXiv:1407.8136 [PDF]
(Submitted on 30 Jul 2014)
Phys. Rev. D90 (2014) 112002
Inspire 1308922
Tools
Abstract
An analysis of B0->D K*0 decays is presented, where D represents an admixture of D0 and D0b mesons reconstructed in four separate final states: K-pi+, pi-K+, K+K- and pi+pi-. The data sample corresponds to 3.0fb-1 of proton-proton collision, collected by the LHCb experiment. Measurements of several observables are performed, including CP asymmetries. The most precise determination is presented of rB(DK*0), the magnitude of the ratio of the amplitudes of the decay B0->D K+ pi- with a b->u or a b->c transition, in a K pi mass region of +/-50 MeV/c2 around the K*(892) mass and for an absolute value of the cosine of the K*0 helicity angle larger than 0.4.
Figures and captions
|
Feynman diagrams of {\it(left)} $ B ^0 \rightarrow D ^0 K ^{*0} $ and {\it (right)} $ B ^0 \rightarrow \overline{ D }{} {}^0 K ^{*0} $. |
feynma[..].pdf [1 MiB] HiDef png [84 KiB] Thumbnail [56 KiB] *.C file |

|
|
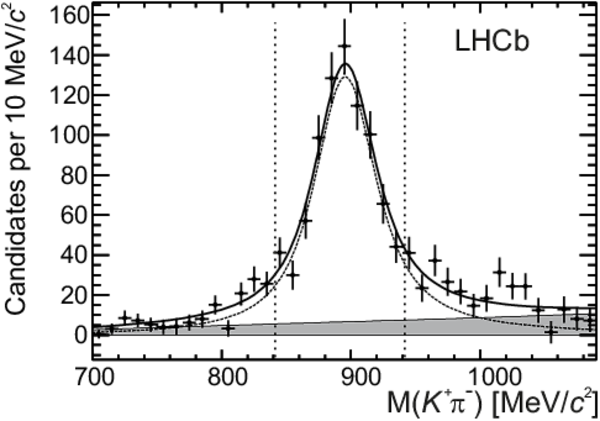
Background-subtracted $ K ^{*0} \rightarrow K ^+ \pi ^- $ invariant mass for $ B ^0 \rightarrow D ( K ^+ \pi ^- ) K ^{*0} $ signal candidates. The data (points) and the fit described in the text (solid line) are shown. The dashed line represents the $ K ^{*0} $ signal and the filled area the non- $ K ^{*0}$ contribution to the $ B ^0 \rightarrow D K ^{*0} $ signal. The vertical dotted lines indicate the invariant mass region used in the analysis. |
kstarmm_1.pdf [1 MiB] HiDef png [218 KiB] Thumbnail [150 KiB] *.C file |
|
|
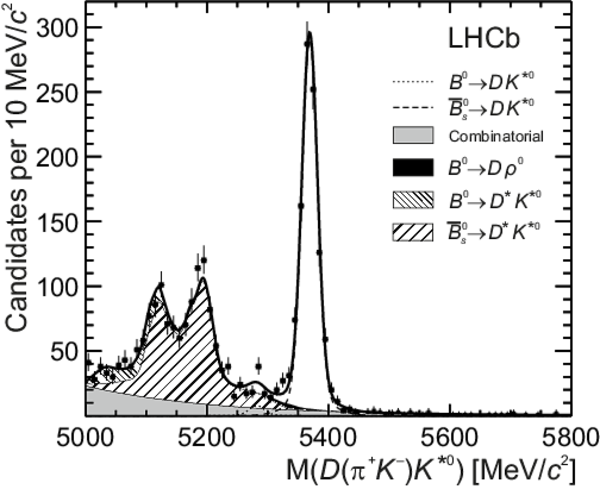
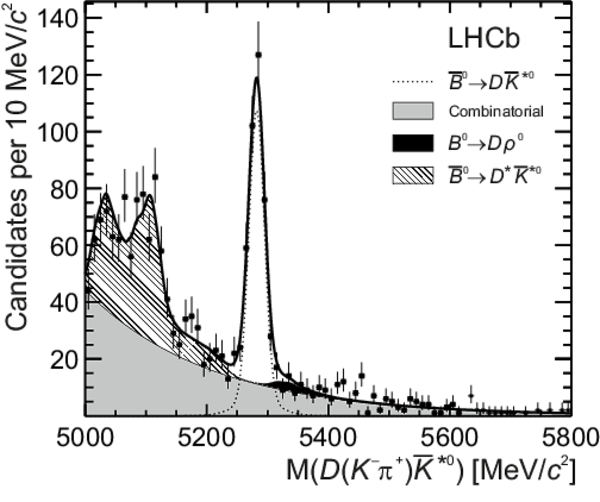
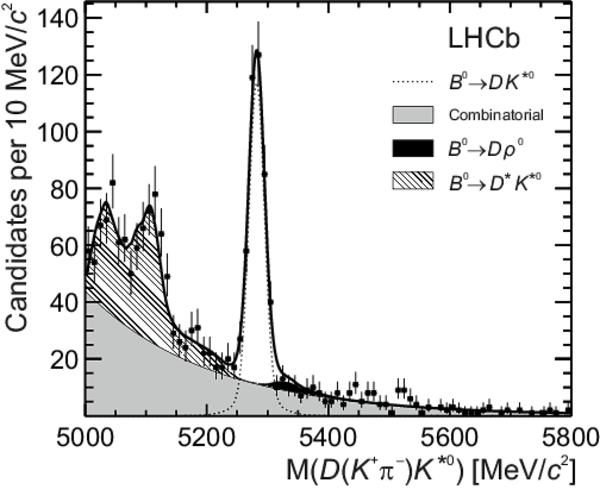
Distributions of (top left) $ D (\pi ^- K ^+ ) \overline{ K }{} {}^{*0} $, (top right) $ D (\pi ^+ K ^- ) K ^{*0} $, (bottom left) $ D ( K ^- \pi ^+ ) \overline{ K }{} {}^{*0} $ and (bottom right) $ D ( K ^+ \pi ^- ) K ^{*0} $ invariant mass. The data (black points) and the fitted invariant mass model (thick solid line) are shown. The PDFs corresponding to the different species are indicated in the legend: the $ B ^0 $ signal, the $ B ^0_ s $ signal, combinatorial background, $ B ^0 \rightarrow D \rho^0$ background, partially reconstructed $ B ^0_ s \rightarrow D ^* \overline{ K }{} {}^{*0} $ and $ B ^0 \rightarrow D ^* K ^{*0} $ backgrounds. |
b0bar_[..].pdf [1 MiB] HiDef png [282 KiB] Thumbnail [228 KiB] *.C file |

|
|
b0_os_[..].pdf [1 MiB] HiDef png [337 KiB] Thumbnail [226 KiB] *.C file |
|
|
|
b0bar_[..].pdf [1 MiB] HiDef png [303 KiB] Thumbnail [247 KiB] *.C file |
|
|
|
b0_ss_[..].pdf [1 MiB] HiDef png [297 KiB] Thumbnail [241 KiB] *.C file |
|
|
|
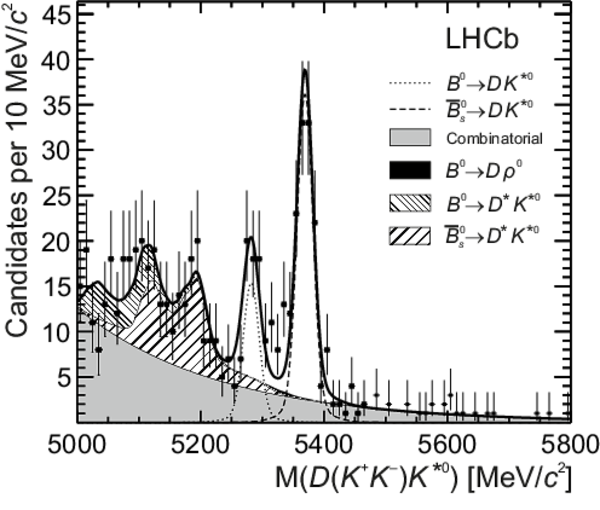
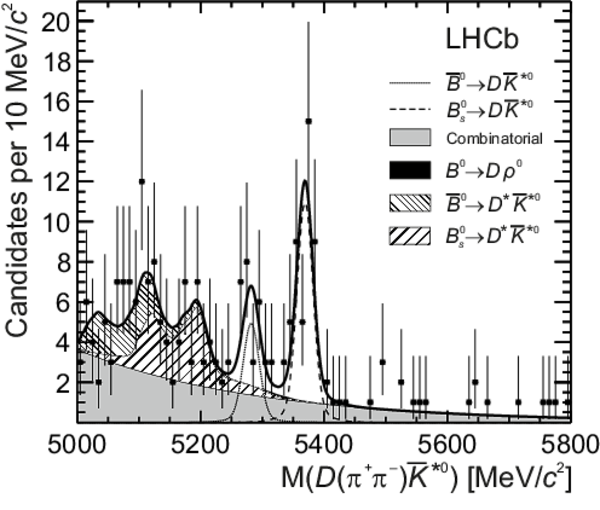
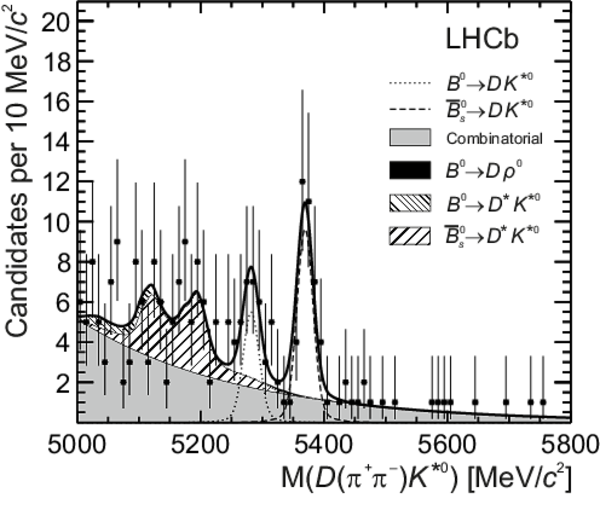
Distributions of (top left) $ D ( K ^+ K ^- ) \overline{ K }{} {}^{*0} $, (top right) $ D ( K ^+ K ^- ) K ^{*0} $, (bottom left) $ D (\pi ^+ \pi ^- ) \overline{ K }{} {}^{*0} $ and (bottom right) $ D (\pi ^+ \pi ^- ) K ^{*0} $ invariant mass. The data (black points) and the fitted invariant mass model (thick solid line) are shown. The PDFs corresponding to the different species are indicated in the legend: the $ B ^0 $ signal, the $ B ^0_ s $ signal, combinatorial background, $ B ^0 \rightarrow D \rho^0$ background and partially reconstructed $ B ^0_ s \rightarrow D ^* \overline{ K }{} {}^{*0} $ and $ B ^0 \rightarrow D ^* K ^{*0} $ backgrounds. |
b0bar_[..].pdf [1 MiB] HiDef png [401 KiB] Thumbnail [287 KiB] *.C file |

|
|
b0_kk_[..].pdf [1 MiB] HiDef png [401 KiB] Thumbnail [287 KiB] *.C file |
|
|
|
b0bar_[..].pdf [1 MiB] HiDef png [385 KiB] Thumbnail [273 KiB] *.C file |
|
|
|
b0_pip[..].pdf [1 MiB] HiDef png [361 KiB] Thumbnail [265 KiB] *.C file |
|
|
|
$p$-value as a function of $r_{ B }$, for $\kappa=0.95\pm0.03$. The horizontal dashed lines represent the $1\sigma$ and $2\sigma$ confidence levels and the vertical dotted line represents the obtained central value. |
rb_scan_1d.pdf [1 MiB] HiDef png [209 KiB] Thumbnail [143 KiB] *.C file |

|
|
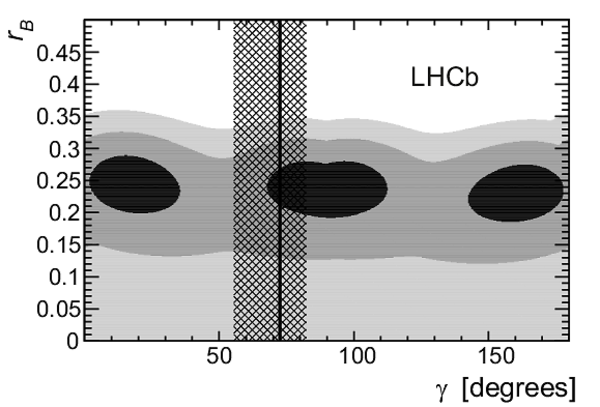
Two-dimensional projections of the $p$-value in $(r_B,\delta_B,\gamma )$ parameter space onto (left) $r_{ B }$ and $\gamma$ and (right) $\delta_{ B }$ and $\gamma$, for $\kappa=0.95\pm0.03$. The contours are the $n\sigma$ profile likelihood contours, where $\Delta\chi^2 = n^2$ with $n = 1$ (black), 2 (medium grey), and 3 (light gray), corresponding to 39.4%, 86.5% and 98.9% confidence level, respectively. The vertical line and hashed band represent the best-fit value of $\gamma $ and the 68.3% confidence level interval by Ref. [3]. |
newcon[..].pdf [1 MiB] HiDef png [390 KiB] Thumbnail [323 KiB] *.C file |
|
|
newcon[..].pdf [1 MiB] HiDef png [443 KiB] Thumbnail [344 KiB] *.C file |

|
|
|
Animated gif made out of all figures. |
PAPER-2014-028.gif Thumbnail |

|
Tables and captions
|
Yields of signal candidates with their statistical uncertainties. |
Table_1.pdf [51 KiB] HiDef png [83 KiB] Thumbnail [39 KiB] tex code |

|
|
Uncertainties in the observables. All model-related systematic uncertainties are added in quadrature and the result is shown as one source of systematic uncertainty. The presence of `--' indicates that the source of uncertainty does not affect the observable. |
Table_2.pdf [55 KiB] HiDef png [118 KiB] Thumbnail [51 KiB] tex code |

|
Supplementary Material [file]
| Supplementary material full pdf |
supple[..].pdf [74 KiB] |

|
Created on 02 May 2024.
