The charmless three-body decays $B^0_{(s)} \to K_{\mathrm{\scriptscriptstyle
S}}^0 h^+ h^{\prime -}$ (where $h^{(\prime)} = \pi, K$) are analysed using a
sample of $pp$ collision data recorded by the LHCb experiment, corresponding to
an integrated luminosity of $3 fb^{-1}$. The branching fractions are
measured relative to that of the $B^0 \to K_{\mathrm{\scriptscriptstyle S}}^0
\pi^{+} \pi^{-}$ decay, and are determined to be: \begin{eqnarray*}
\frac{{\cal B}(B^0\rightarrow K^0_{\rm S}K^{\pm}\pi^{\mp})}{{\cal
B}(B^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})}
= {} & 0.123 \pm 0.009 \; \mathrm{ (stat)}\; \pm 0.015 \;
\mathrm{ (syst)} ,
\frac{{\cal B}(B^0\rightarrow K^0_{\rm S}K^{+}K^{-})} {{\cal
B}(B^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})}
= {} & 0.549 \pm 0.018 \; \mathrm{ (stat)}\; \pm 0.033 \;
\mathrm{ (syst)} ,
\frac{{\cal B}(B_{s}^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})}{{\cal
B}(B^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})}
= {} & 0.191 \pm 0.027 \; \mathrm{ (stat)}\; \pm 0.031 \;
\mathrm{ (syst)}\; \pm 0.011 \; (f_s/f_d) ,
\frac{{\cal B}(B_{s}^0\rightarrow K^0_{\rm S}K^{\pm}\pi^{\mp})} {{\cal
B}(B^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})}
= {} & 1.70\phantom{0} \pm 0.07\phantom{0} \; \mathrm{ (stat)} \; \pm
0.11\phantom{0} \; \mathrm{ (syst)}\; \pm 0.10\phantom{0} \; (f_s/f_d) ,
\frac{{\cal B}(B_{s}^0\rightarrow K^0_{\rm S}K^{+}K^{-})}{{\cal
B}(B^0\rightarrow K^0_{\rm S}\pi^{+}\pi^{-})} \in {} & [0.008 - 0.051] \rm
at 90\% confidence level,
\end{eqnarray*} where $f_s/f_d$ represents the ratio of hadronisation
fractions of the $B^0_s$ and $B^0$ mesons.
|
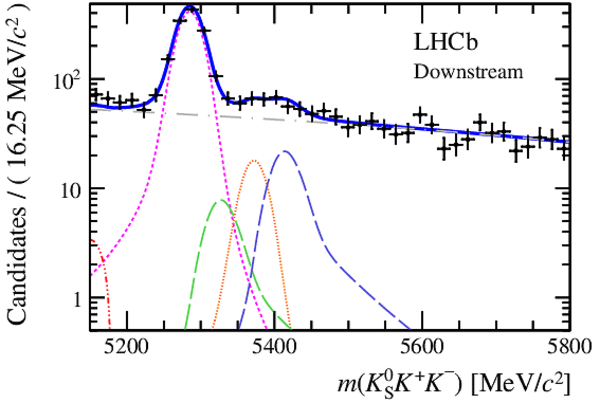
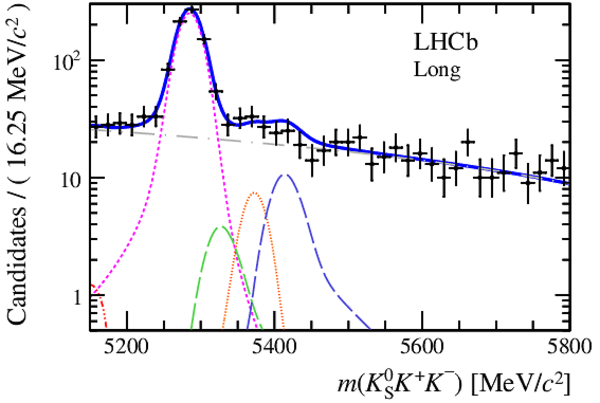
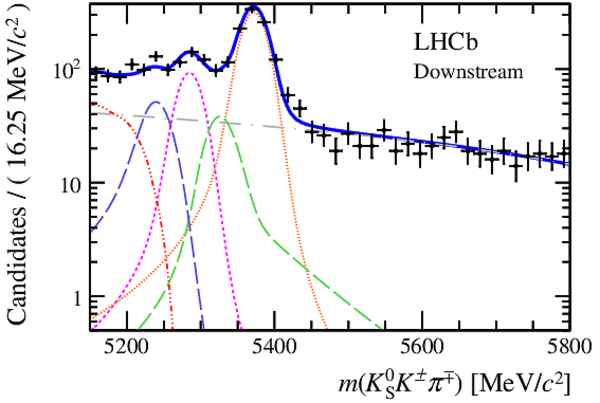
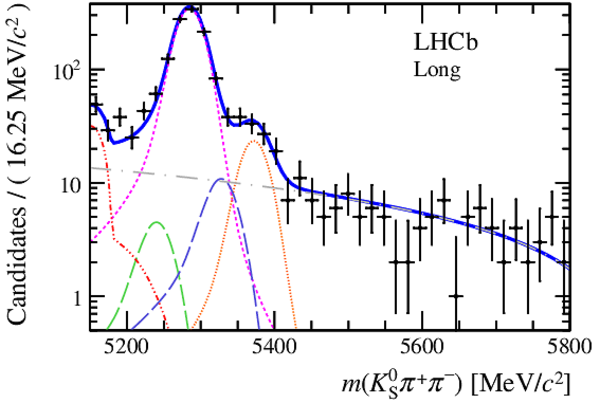
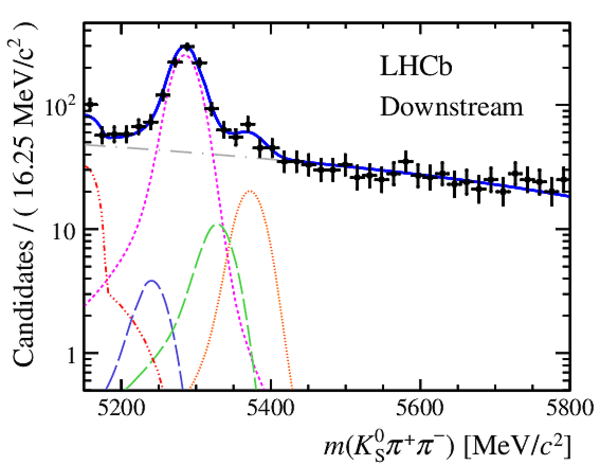
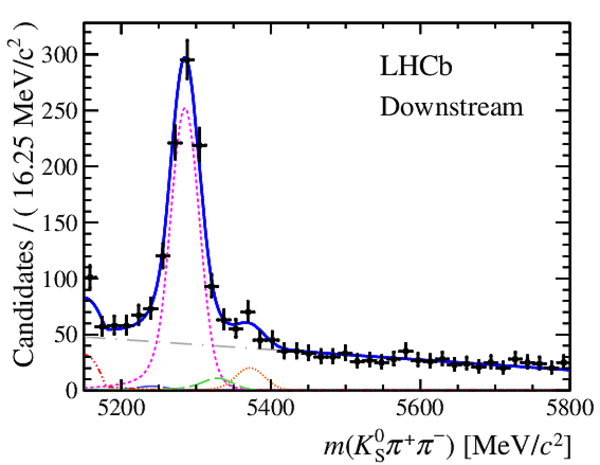
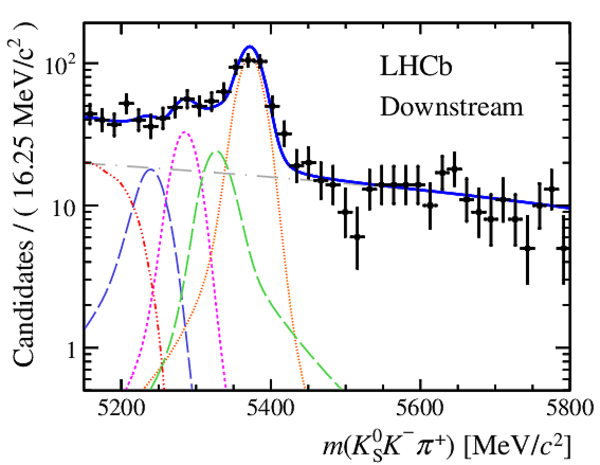
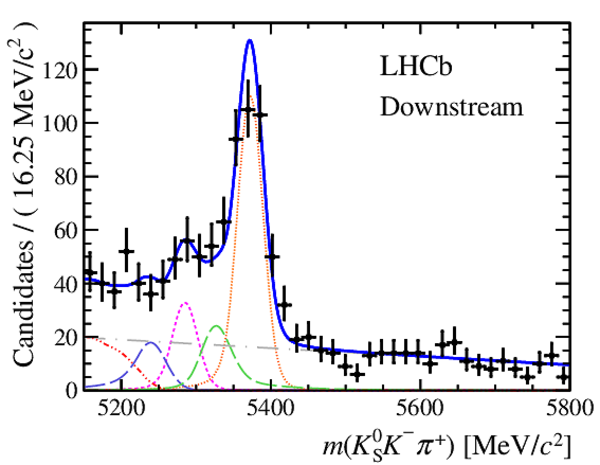
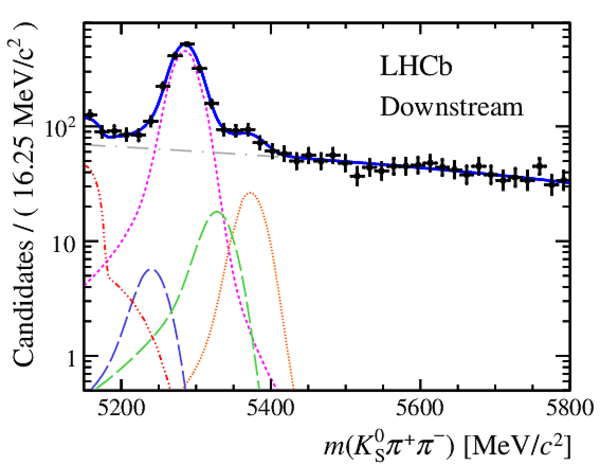
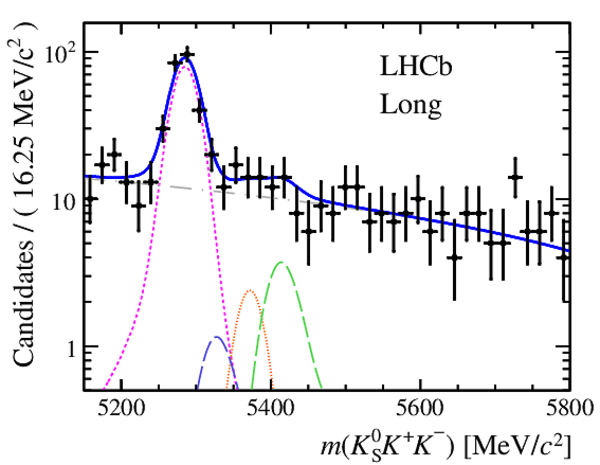
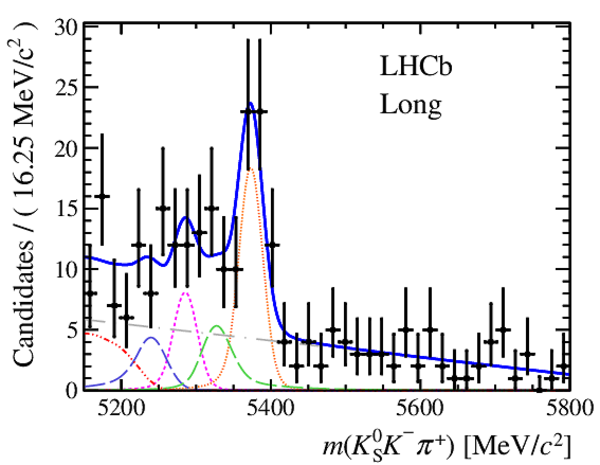
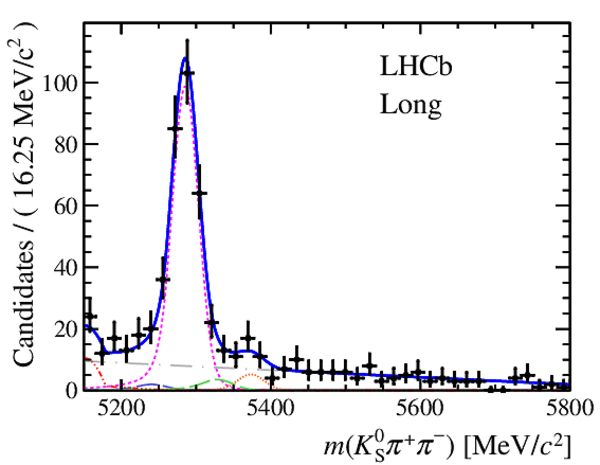
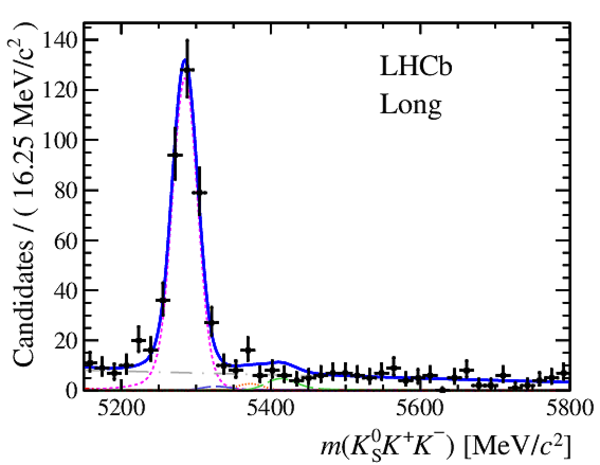
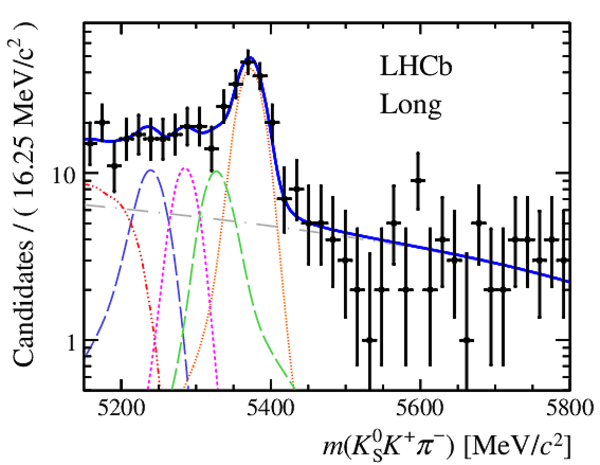
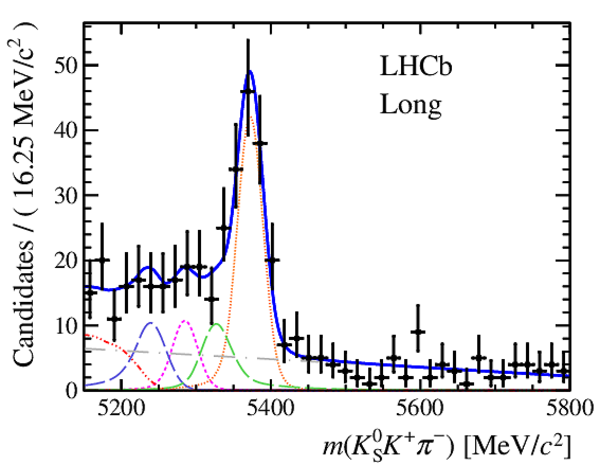
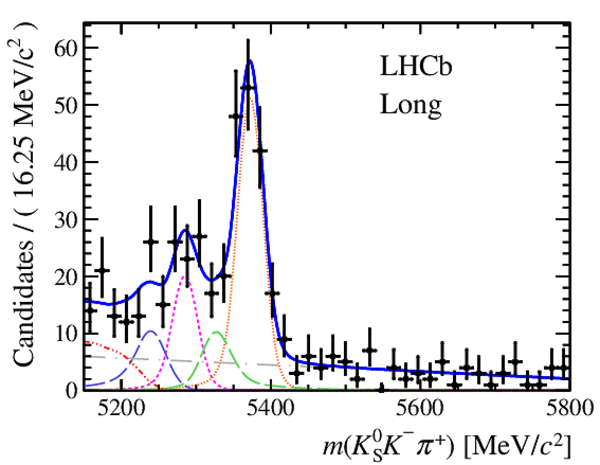
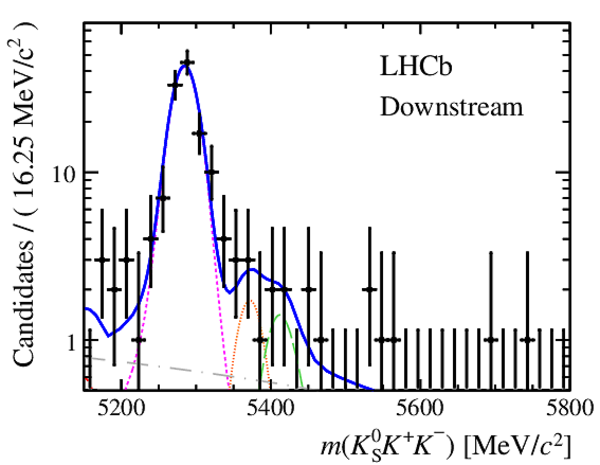
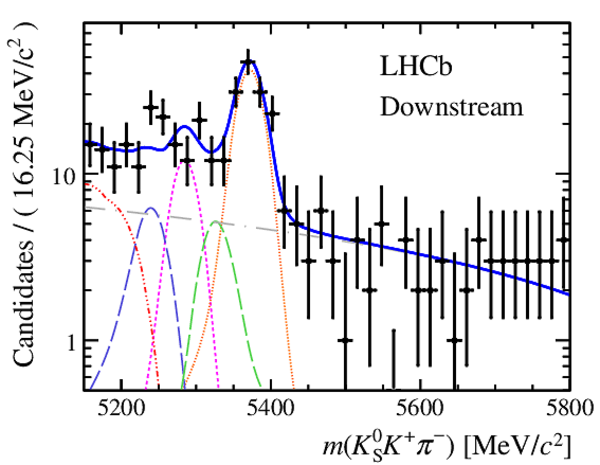
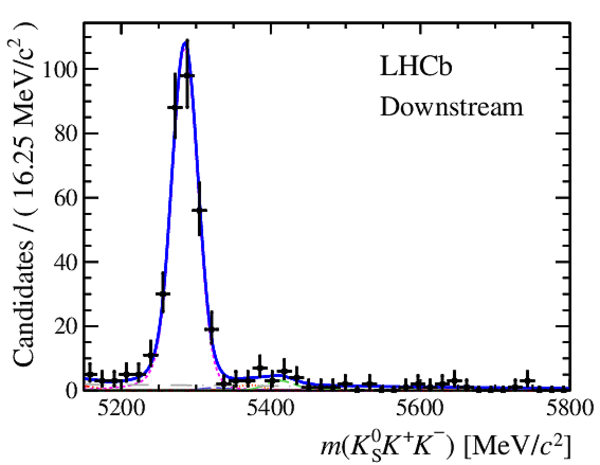
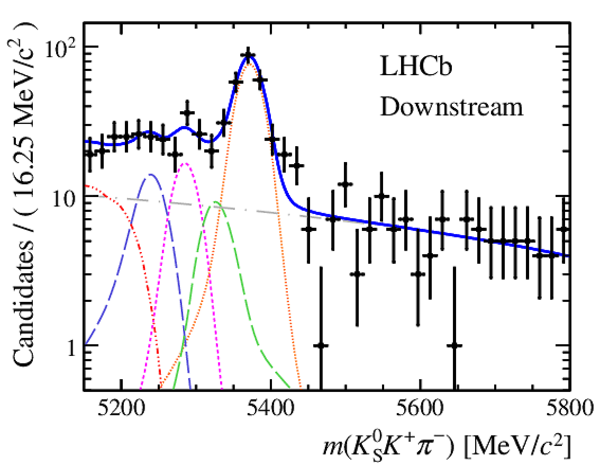
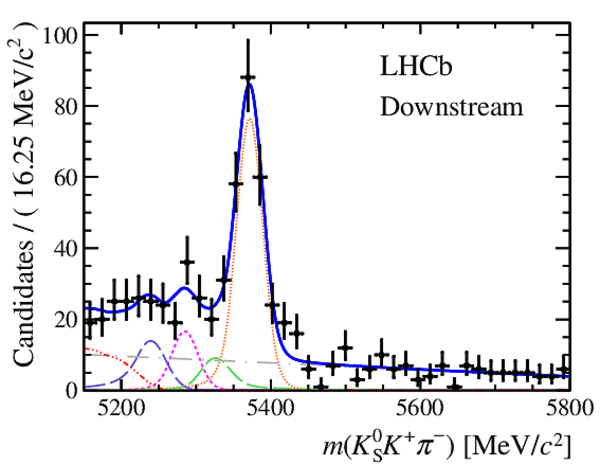
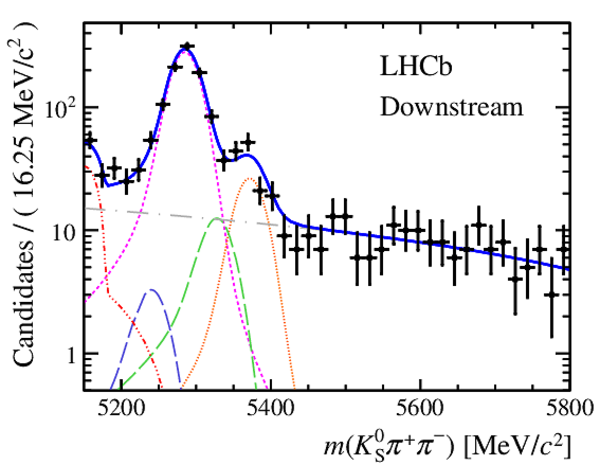
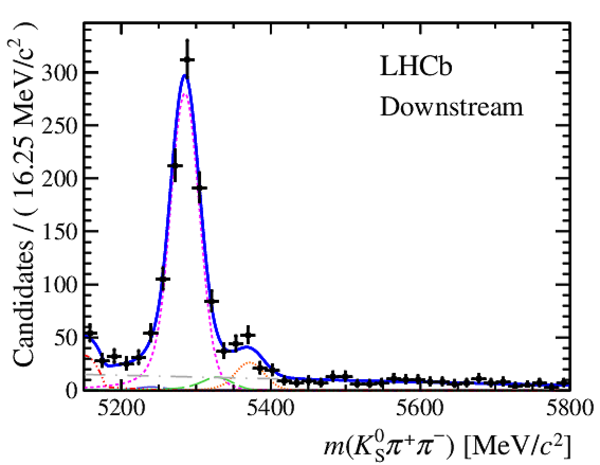
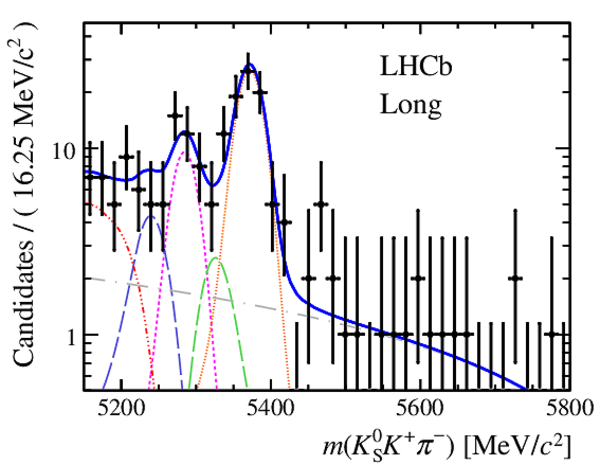
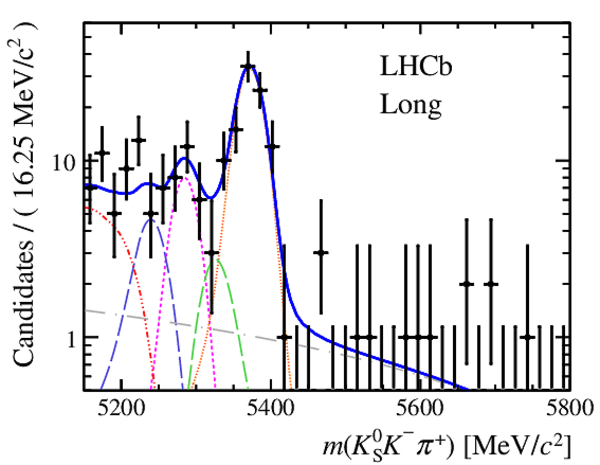
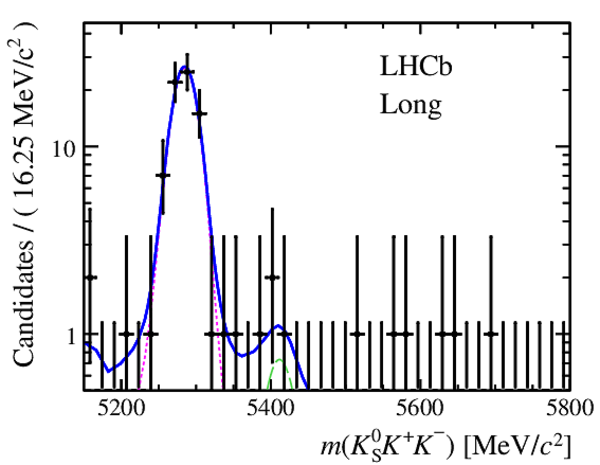
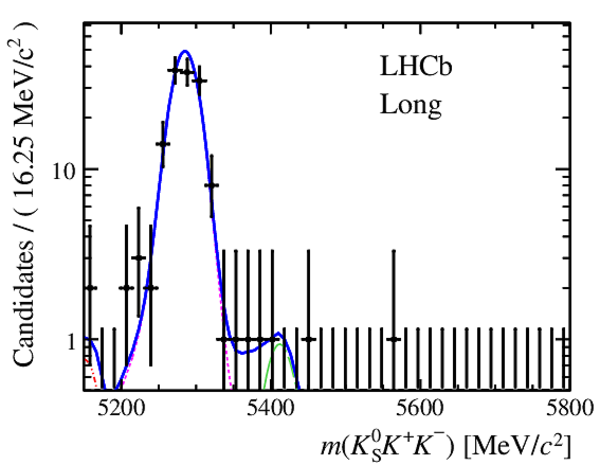
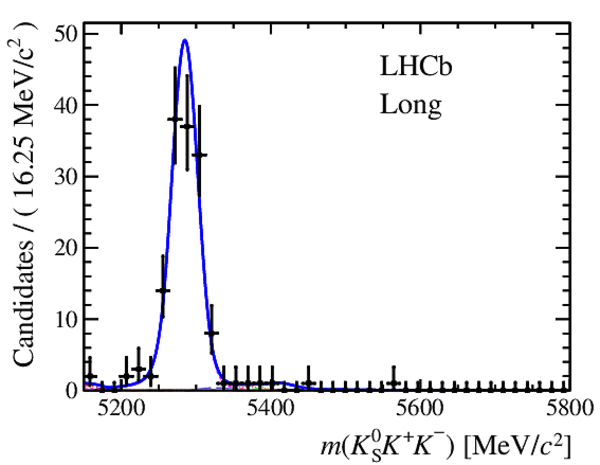
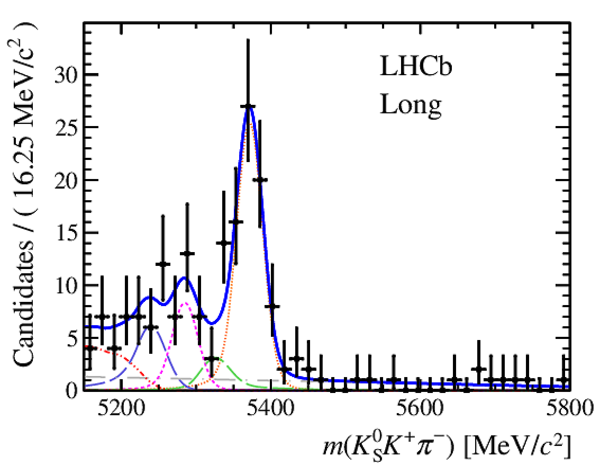
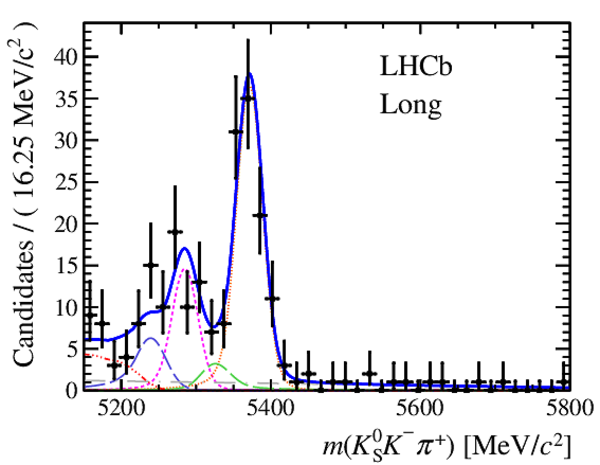
Invariant mass distributions of, from top to bottom, $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^\pm \pi ^\mp $
and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ candidates, summing the three periods of
data taking, with the selection optimisation chosen
for the favoured decay modes for (left) downstream and
(right) long $ K ^0_{\mathrm{ \scriptscriptstyle S}}$ reconstruction categories.
In each plot, data are the black points with error bars and the total fit model is overlaid (solid blue line).
The $ B ^0$ ( $ B ^0_ s $ ) signal components are the pink (orange) short-dashed (dotted) lines,
while fully reconstructed misidentified decays are the green and dark blue dashed lines
close to the $ B ^0$ and $ B ^0_ s $ peaks.
The sum of the partially reconstructed contributions from $ B $ to open charm decays,
charmless hadronic decays, $ B ^0 \rightarrow \eta ^{\prime} K ^0_{\mathrm{ \scriptscriptstyle S}} $ and charmless radiative decays are
the red dash triple-dotted lines.
The combinatorial background contribution is the gray long-dash dotted
line.
|
FusedC[..].pdf [21 KiB]
HiDef png [246 KiB]
Thumbnail [185 KiB]
*.C file
|
|
FusedC[..].pdf [21 KiB]
HiDef png [235 KiB]
Thumbnail [177 KiB]
*.C file
|
|
FusedC[..].pdf [21 KiB]
HiDef png [276 KiB]
Thumbnail [198 KiB]
*.C file
|

|
FusedC[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [199 KiB]
*.C file
|

|
FusedC[..].pdf [21 KiB]
HiDef png [248 KiB]
Thumbnail [183 KiB]
*.C file
|
|
FusedC[..].pdf [21 KiB]
HiDef png [246 KiB]
Thumbnail [183 KiB]
*.C file
|
|
|
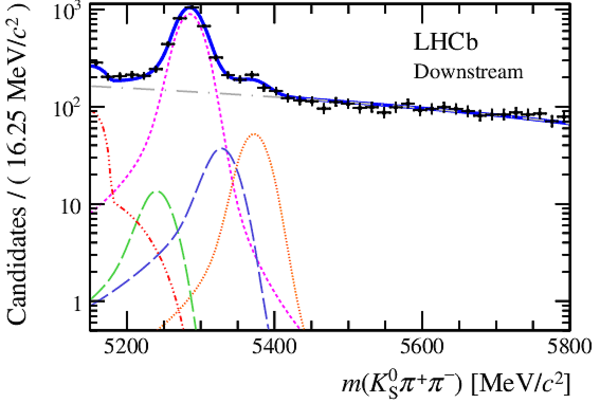
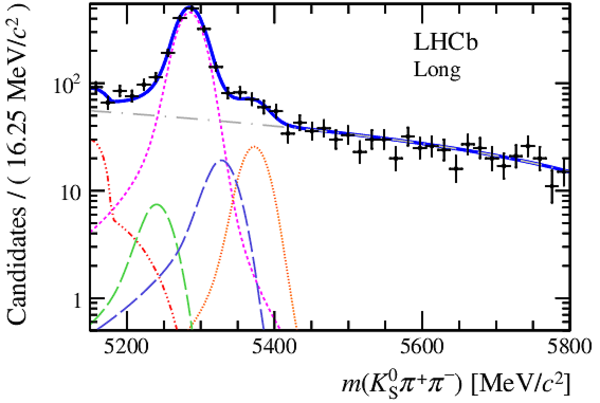
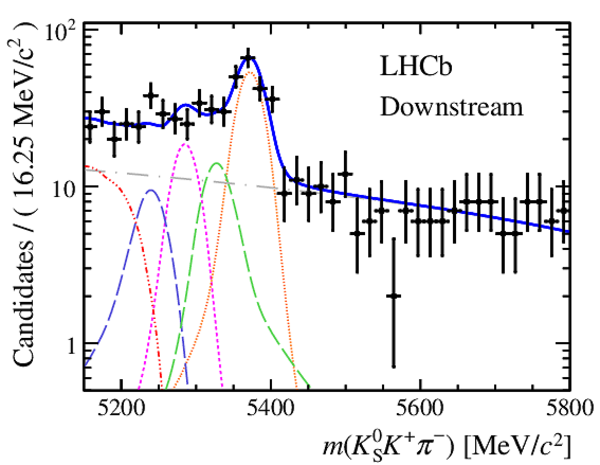
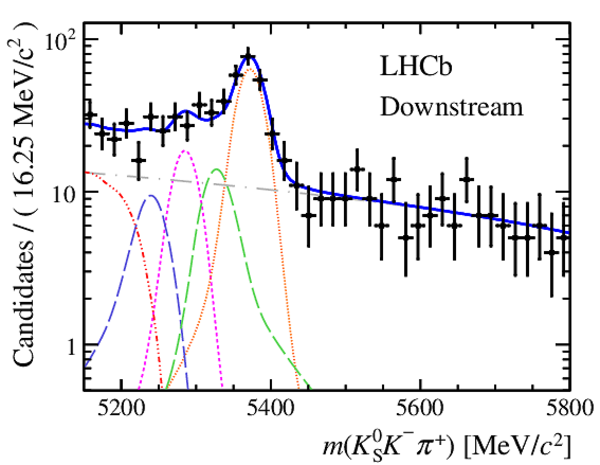
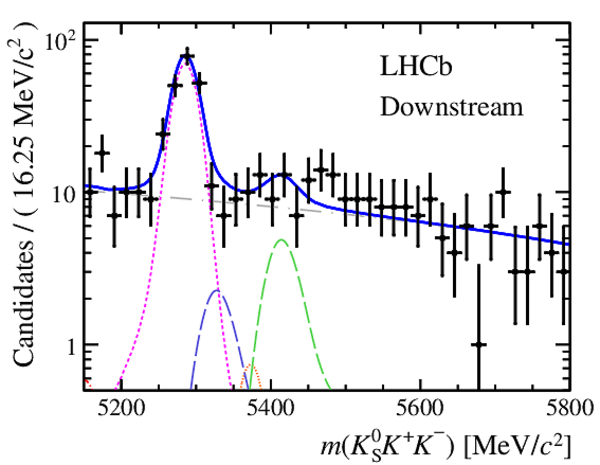
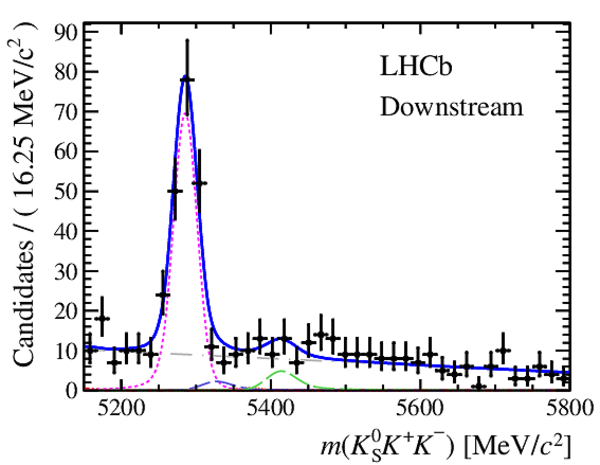
Invariant mass distributions of, from top to bottom, $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^\pm \pi ^\mp $
and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ candidates, summing the three periods of
data taking, with the selection optimisation chosen
for the favoured decay modes for (left) downstream and
(right) long $ K ^0_{\mathrm{ \scriptscriptstyle S}}$ reconstruction categories.
In each plot, data are the black points with error bars and the total fit model is overlaid (solid blue line).
The $ B ^0$ ( $ B ^0_ s $ ) signal components are the pink (orange) short-dashed (dotted) lines,
while fully reconstructed misidentified decays are the green and dark blue dashed lines
close to the $ B ^0$ and $ B ^0_ s $ peaks.
The sum of the partially reconstructed contributions from $ B $ to open charm decays,
charmless hadronic decays, $ B ^0 \rightarrow \eta ^{\prime} K ^0_{\mathrm{ \scriptscriptstyle S}} $ and charmless radiative decays are
the red dash triple-dotted lines.
The combinatorial background contribution is the gray long-dash dotted
line.
|
FusedC[..].pdf [20 KiB]
HiDef png [244 KiB]
Thumbnail [184 KiB]
*.C file
|

|
FusedC[..].pdf [19 KiB]
HiDef png [214 KiB]
Thumbnail [166 KiB]
*.C file
|

|
FusedC[..].pdf [21 KiB]
HiDef png [276 KiB]
Thumbnail [201 KiB]
*.C file
|
|
FusedC[..].pdf [21 KiB]
HiDef png [276 KiB]
Thumbnail [199 KiB]
*.C file
|

|
FusedC[..].pdf [21 KiB]
HiDef png [255 KiB]
Thumbnail [190 KiB]
*.C file
|

|
FusedC[..].pdf [21 KiB]
HiDef png [254 KiB]
Thumbnail [187 KiB]
*.C file
|
|
|
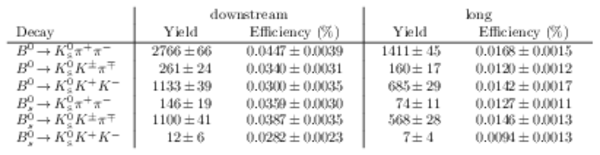
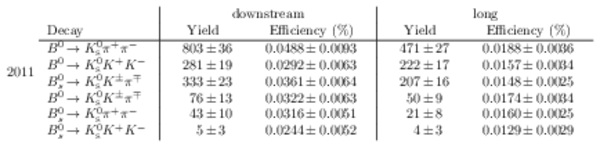
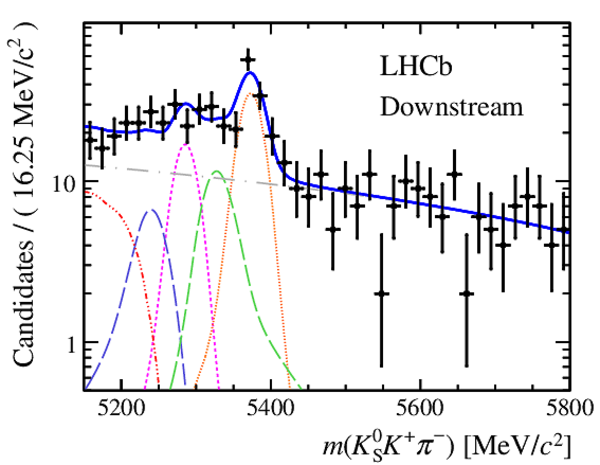
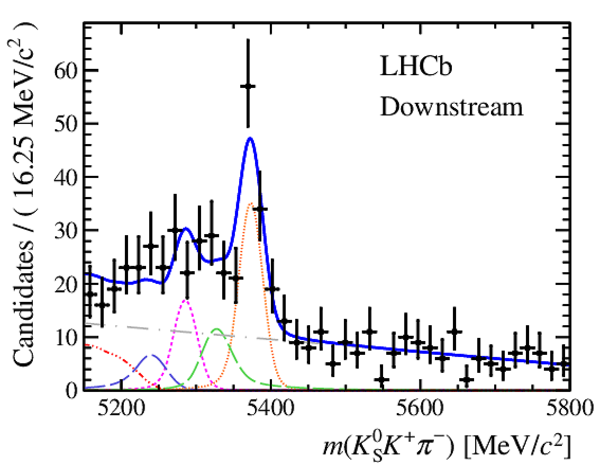
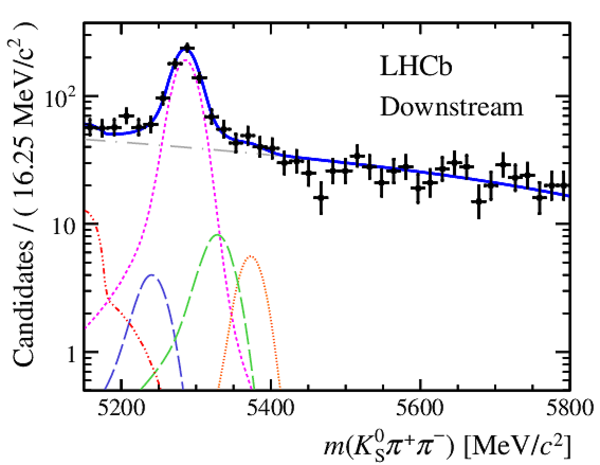
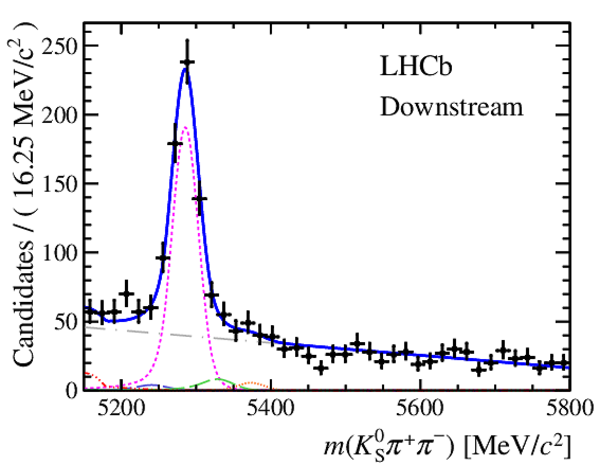
Results of the simultaneous fit to data (downstream, 2011) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [20 KiB]
HiDef png [252 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [252 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [254 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [254 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [286 KiB]
Thumbnail [221 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [286 KiB]
Thumbnail [221 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [282 KiB]
Thumbnail [227 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [282 KiB]
Thumbnail [227 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [219 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [284 KiB]
Thumbnail [233 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [284 KiB]
Thumbnail [233 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [251 KiB]
Thumbnail [203 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [251 KiB]
Thumbnail [203 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [251 KiB]
Thumbnail [206 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [251 KiB]
Thumbnail [206 KiB]
*.C file
|

|
|
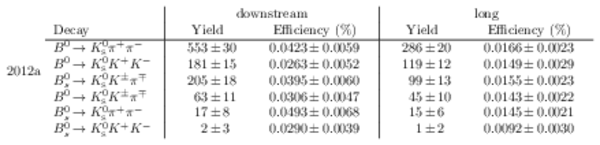
Results of the simultaneous fit to data (downstream, 2012a) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [19 KiB]
HiDef png [244 KiB]
Thumbnail [198 KiB]
*.C file
|
|
from51[..].pdf [19 KiB]
HiDef png [244 KiB]
Thumbnail [198 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [264 KiB]
Thumbnail [219 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [264 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [218 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [218 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [273 KiB]
Thumbnail [220 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [273 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [281 KiB]
Thumbnail [224 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [281 KiB]
Thumbnail [224 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [245 KiB]
Thumbnail [200 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [245 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [246 KiB]
Thumbnail [203 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [246 KiB]
Thumbnail [203 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2012b) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [20 KiB]
HiDef png [249 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [249 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [250 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [250 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [218 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [218 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [218 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [218 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [283 KiB]
Thumbnail [220 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [283 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [252 KiB]
Thumbnail [203 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [252 KiB]
Thumbnail [203 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [244 KiB]
Thumbnail [197 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [244 KiB]
Thumbnail [197 KiB]
*.C file
|

|
|
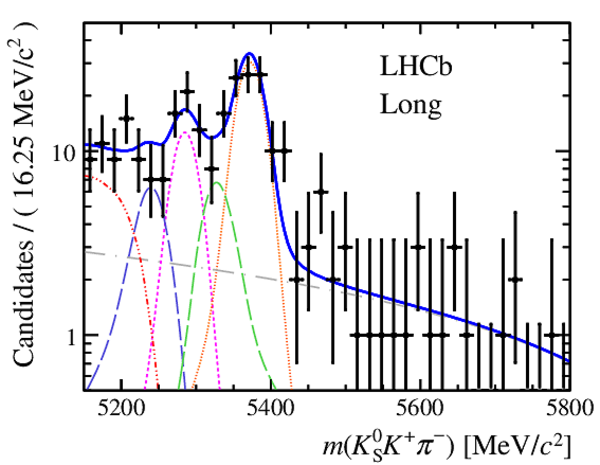
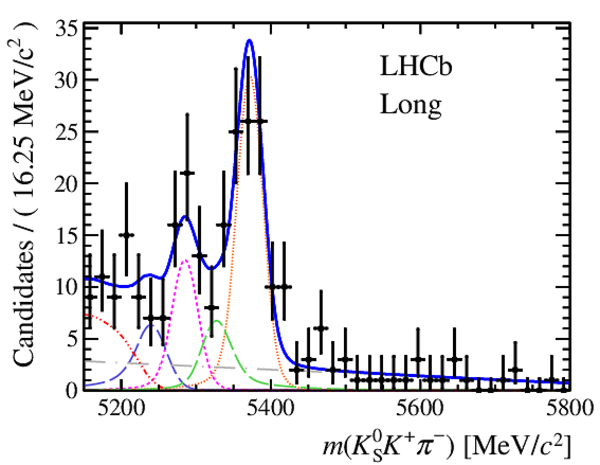
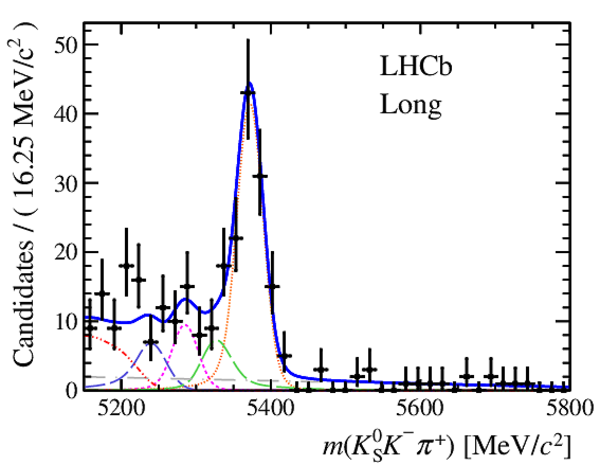
Results of the simultaneous fit to data (downstream, 2011) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [19 KiB]
HiDef png [229 KiB]
Thumbnail [189 KiB]
*.C file
|
|
from51[..].pdf [19 KiB]
HiDef png [229 KiB]
Thumbnail [189 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [237 KiB]
Thumbnail [191 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [237 KiB]
Thumbnail [191 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [290 KiB]
Thumbnail [210 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [290 KiB]
Thumbnail [210 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [285 KiB]
Thumbnail [219 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [285 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [280 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [280 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [251 KiB]
Thumbnail [193 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [251 KiB]
Thumbnail [193 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [247 KiB]
Thumbnail [201 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [247 KiB]
Thumbnail [201 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [253 KiB]
Thumbnail [209 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [253 KiB]
Thumbnail [209 KiB]
*.C file
|

|
|
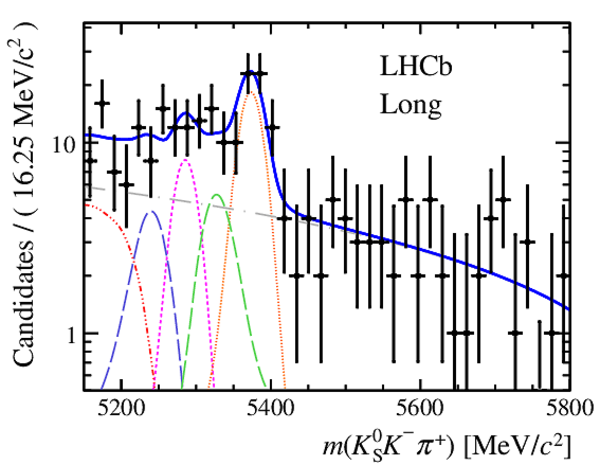
Results of the simultaneous fit to data (downstream, 2012a) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [19 KiB]
HiDef png [238 KiB]
Thumbnail [188 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [238 KiB]
Thumbnail [188 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [237 KiB]
Thumbnail [189 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [237 KiB]
Thumbnail [189 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [208 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [208 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [280 KiB]
Thumbnail [213 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [280 KiB]
Thumbnail [213 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [280 KiB]
Thumbnail [208 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [280 KiB]
Thumbnail [208 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [215 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [215 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [245 KiB]
Thumbnail [197 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [245 KiB]
Thumbnail [197 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [237 KiB]
Thumbnail [188 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [237 KiB]
Thumbnail [188 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2012b) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [20 KiB]
HiDef png [239 KiB]
Thumbnail [193 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [239 KiB]
Thumbnail [193 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [237 KiB]
Thumbnail [190 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [237 KiB]
Thumbnail [190 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [212 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [212 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [212 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [212 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [289 KiB]
Thumbnail [217 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [289 KiB]
Thumbnail [217 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [281 KiB]
Thumbnail [219 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [281 KiB]
Thumbnail [219 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [246 KiB]
Thumbnail [199 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [246 KiB]
Thumbnail [199 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [238 KiB]
Thumbnail [191 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [238 KiB]
Thumbnail [191 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2011) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [18 KiB]
HiDef png [213 KiB]
Thumbnail [171 KiB]
*.C file
|
|
from51[..].pdf [18 KiB]
HiDef png [213 KiB]
Thumbnail [171 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [209 KiB]
Thumbnail [172 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [209 KiB]
Thumbnail [172 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [214 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [214 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [271 KiB]
Thumbnail [213 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [271 KiB]
Thumbnail [213 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [283 KiB]
Thumbnail [215 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [283 KiB]
Thumbnail [215 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [217 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [275 KiB]
Thumbnail [217 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [259 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [259 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [247 KiB]
Thumbnail [204 KiB]
*.C file
|
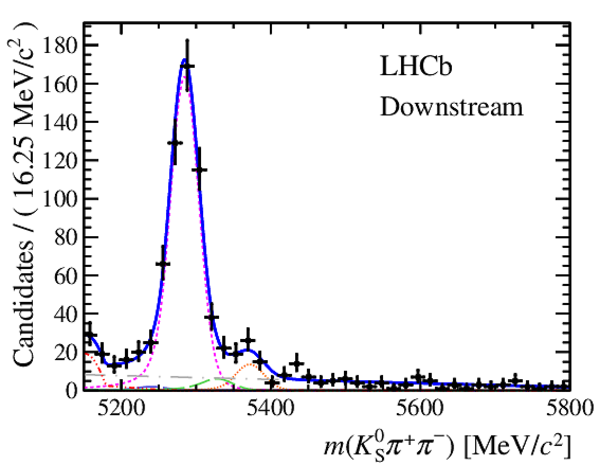
|
from51[..].pdf [21 KiB]
HiDef png [247 KiB]
Thumbnail [204 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2012a) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [19 KiB]
HiDef png [233 KiB]
Thumbnail [186 KiB]
*.C file
|
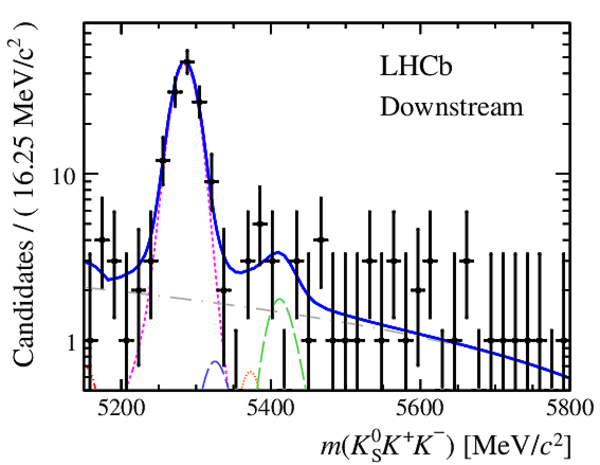
|
from51[..].pdf [19 KiB]
HiDef png [233 KiB]
Thumbnail [186 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [228 KiB]
Thumbnail [184 KiB]
*.C file
|
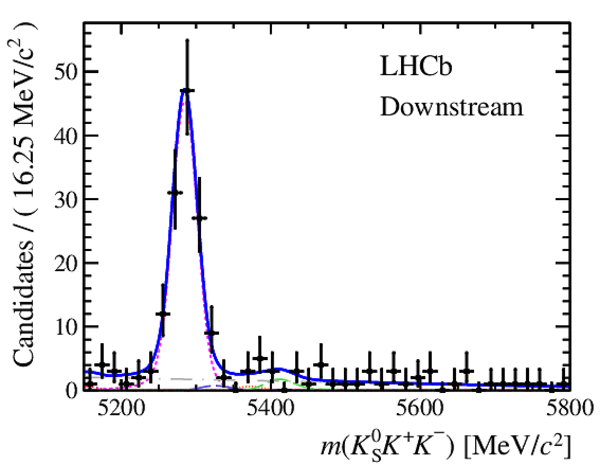
|
from51[..].pdf [20 KiB]
HiDef png [228 KiB]
Thumbnail [184 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [286 KiB]
Thumbnail [214 KiB]
*.C file
|
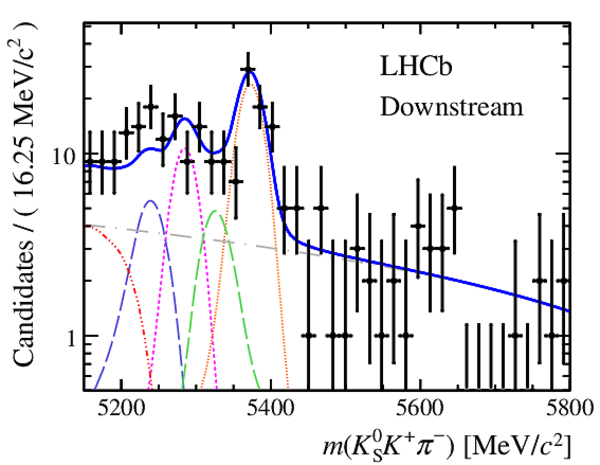
|
from51[..].pdf [20 KiB]
HiDef png [286 KiB]
Thumbnail [214 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [222 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [222 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [211 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [282 KiB]
Thumbnail [211 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [222 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [222 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [254 KiB]
Thumbnail [207 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [254 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [243 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [243 KiB]
Thumbnail [200 KiB]
*.C file
|

|
|
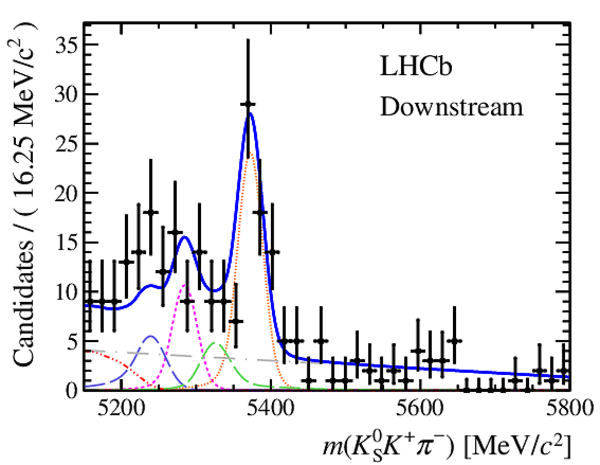
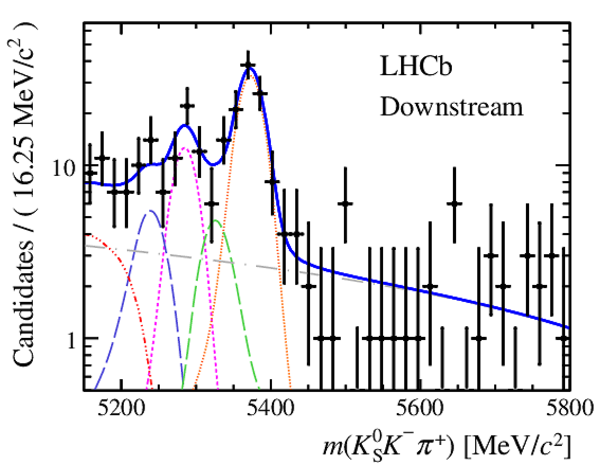
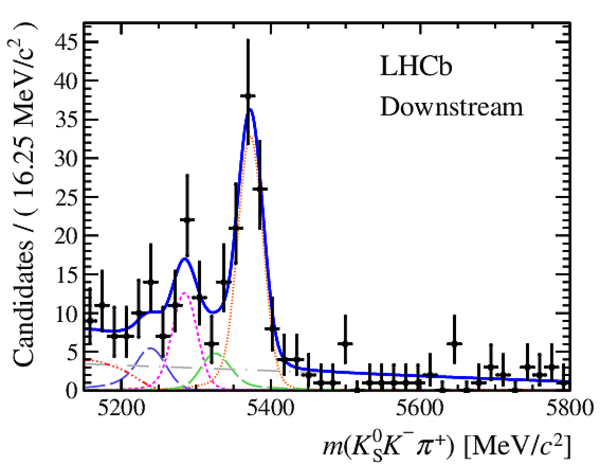
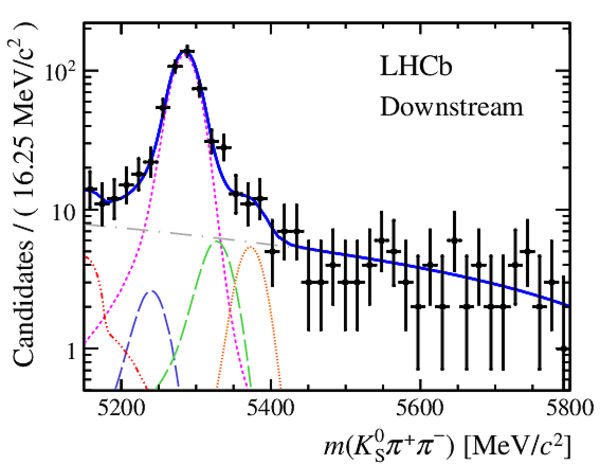
Results of the simultaneous fit to data (downstream, 2012b) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [19 KiB]
HiDef png [239 KiB]
Thumbnail [194 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [239 KiB]
Thumbnail [194 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [216 KiB]
Thumbnail [171 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [216 KiB]
Thumbnail [171 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [218 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [284 KiB]
Thumbnail [218 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [267 KiB]
Thumbnail [207 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [267 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [287 KiB]
Thumbnail [220 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [278 KiB]
Thumbnail [212 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [278 KiB]
Thumbnail [212 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [262 KiB]
Thumbnail [211 KiB]
*.C file
|
|
from51[..].pdf [20 KiB]
HiDef png [262 KiB]
Thumbnail [211 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [242 KiB]
Thumbnail [199 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [242 KiB]
Thumbnail [199 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2011) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [17 KiB]
HiDef png [190 KiB]
Thumbnail [155 KiB]
*.C file
|

|
from51[..].pdf [17 KiB]
HiDef png [190 KiB]
Thumbnail [155 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [193 KiB]
Thumbnail [164 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [193 KiB]
Thumbnail [164 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [272 KiB]
Thumbnail [200 KiB]
*.C file
|
|
from51[..].pdf [19 KiB]
HiDef png [272 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [259 KiB]
Thumbnail [203 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [259 KiB]
Thumbnail [203 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [263 KiB]
Thumbnail [195 KiB]
*.C file
|
|
from51[..].pdf [19 KiB]
HiDef png [263 KiB]
Thumbnail [195 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [248 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [248 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [250 KiB]
Thumbnail [199 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [250 KiB]
Thumbnail [199 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [232 KiB]
Thumbnail [185 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [232 KiB]
Thumbnail [185 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2012a) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [17 KiB]
HiDef png [186 KiB]
Thumbnail [148 KiB]
*.C file
|
|
from51[..].pdf [17 KiB]
HiDef png [186 KiB]
Thumbnail [148 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [203 KiB]
Thumbnail [169 KiB]
*.C file
|
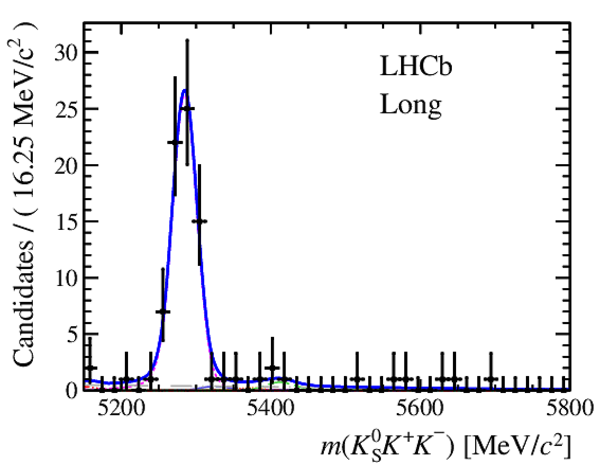
|
from51[..].pdf [20 KiB]
HiDef png [203 KiB]
Thumbnail [169 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [277 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [277 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [284 KiB]
Thumbnail [222 KiB]
*.C file
|
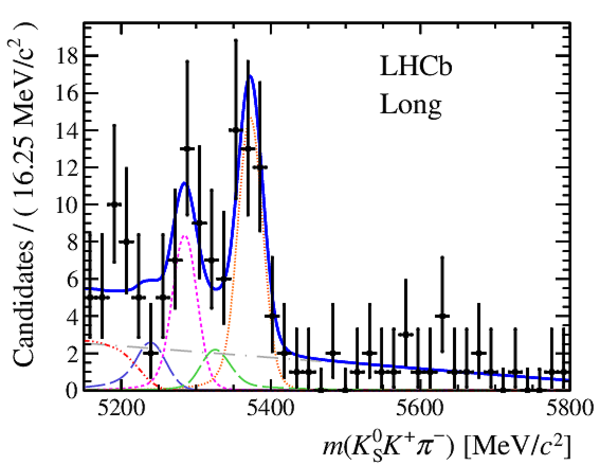
|
from51[..].pdf [21 KiB]
HiDef png [284 KiB]
Thumbnail [222 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [270 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [270 KiB]
Thumbnail [202 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [227 KiB]
*.C file
|
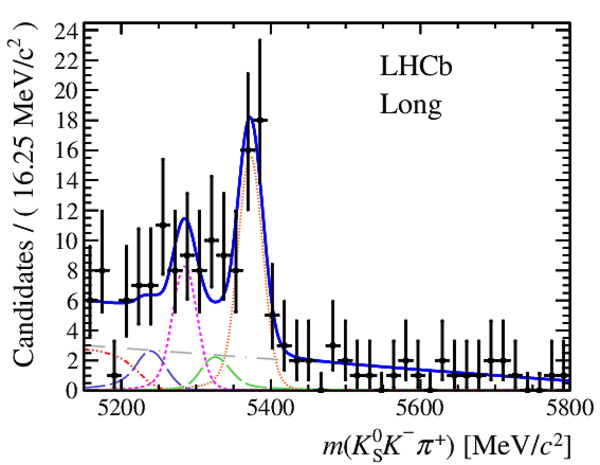
|
from51[..].pdf [21 KiB]
HiDef png [277 KiB]
Thumbnail [227 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [243 KiB]
Thumbnail [192 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [243 KiB]
Thumbnail [192 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [229 KiB]
Thumbnail [194 KiB]
*.C file
|
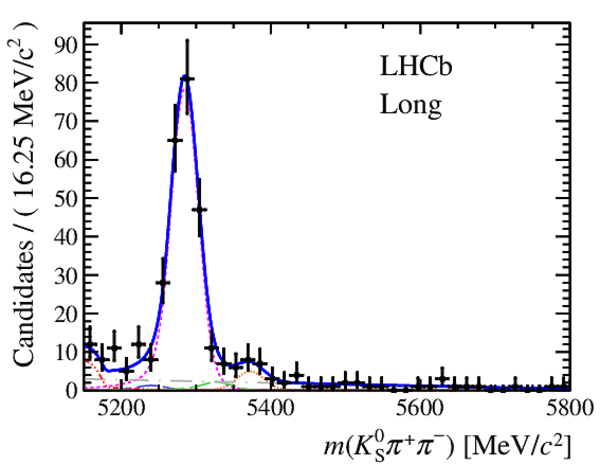
|
from51[..].pdf [21 KiB]
HiDef png [229 KiB]
Thumbnail [194 KiB]
*.C file
|

|
|
Results of the simultaneous fit to data (downstream, 2012b) with the BDT optimisation corresponding to the favoured decay modes. The modes $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ are shown from top to bottom. The left-hand side plots show the results with a logarithmic scale and the right-hand side with a linear scale. Legend is similar to that of plots shown in Fig. ???.
|
from51[..].pdf [17 KiB]
HiDef png [182 KiB]
Thumbnail [146 KiB]
*.C file
|
|
from51[..].pdf [17 KiB]
HiDef png [182 KiB]
Thumbnail [146 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [189 KiB]
Thumbnail [158 KiB]
*.C file
|
|
from51[..].pdf [19 KiB]
HiDef png [189 KiB]
Thumbnail [158 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [271 KiB]
Thumbnail [198 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [271 KiB]
Thumbnail [198 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [253 KiB]
Thumbnail [199 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [253 KiB]
Thumbnail [199 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [269 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [19 KiB]
HiDef png [269 KiB]
Thumbnail [200 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [260 KiB]
Thumbnail [207 KiB]
*.C file
|
|
from51[..].pdf [21 KiB]
HiDef png [260 KiB]
Thumbnail [207 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [248 KiB]
Thumbnail [197 KiB]
*.C file
|

|
from51[..].pdf [20 KiB]
HiDef png [248 KiB]
Thumbnail [197 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [223 KiB]
Thumbnail [184 KiB]
*.C file
|

|
from51[..].pdf [21 KiB]
HiDef png [223 KiB]
Thumbnail [184 KiB]
*.C file
|

|
|
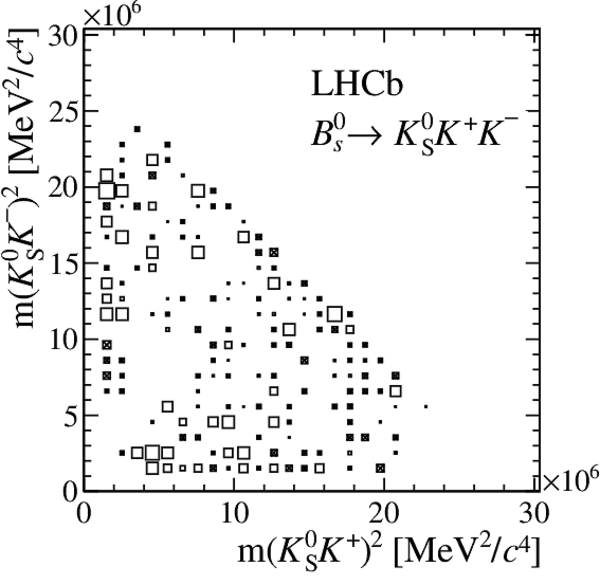
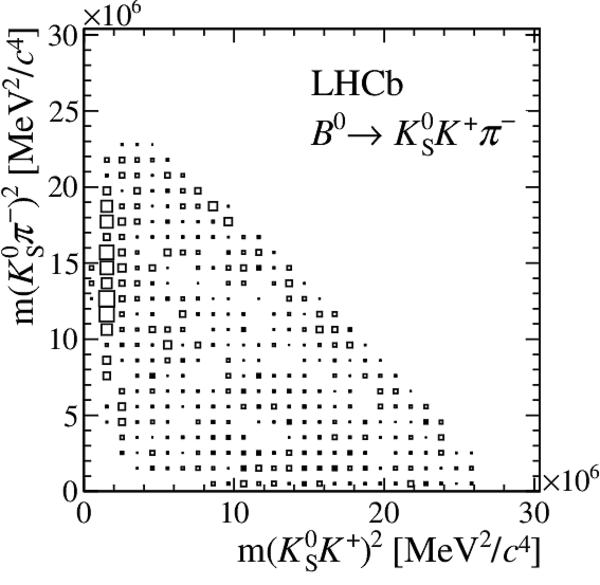
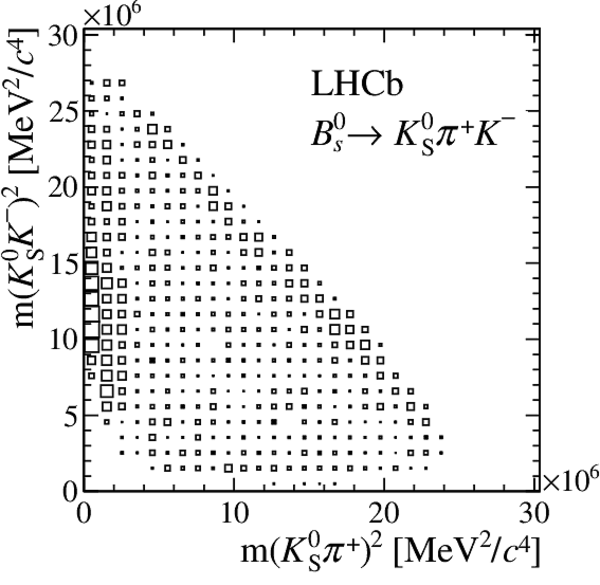
Distribution of sPlot weights in data.
The $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ K ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} K ^+ \pi ^- $ , $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ K ^- $ , and $ K ^0_{\mathrm{ \scriptscriptstyle S}} \pi ^+ \pi ^- $ final states are shown from top to bottom, with a $ B ^0$ parent on the left, and a $ B ^0_ s $ on the right.
All data-taking periods and $ K ^0_{\mathrm{ \scriptscriptstyle S}}$ reconstruction categories are added.
|
Dalitz[..].pdf [19 KiB]
HiDef png [166 KiB]
Thumbnail [108 KiB]
*.C file
|

|
Dalitz[..].pdf [19 KiB]
HiDef png [166 KiB]
Thumbnail [108 KiB]
*.C file
|

|
Dalitz[..].pdf [18 KiB]
HiDef png [146 KiB]
Thumbnail [95 KiB]
*.C file
|
|
Dalitz[..].pdf [18 KiB]
HiDef png [146 KiB]
Thumbnail [95 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [162 KiB]
Thumbnail [105 KiB]
*.C file
|
|
Dalitz[..].pdf [21 KiB]
HiDef png [162 KiB]
Thumbnail [105 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [177 KiB]
Thumbnail [124 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [177 KiB]
Thumbnail [124 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [169 KiB]
Thumbnail [117 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [169 KiB]
Thumbnail [117 KiB]
*.C file
|

|
Dalitz[..].pdf [21 KiB]
HiDef png [177 KiB]
Thumbnail [121 KiB]
*.C file
|
|
Dalitz[..].pdf [21 KiB]
HiDef png [177 KiB]
Thumbnail [121 KiB]
*.C file
|

|
Dalitz[..].pdf [20 KiB]
HiDef png [181 KiB]
Thumbnail [127 KiB]
*.C file
|

|
Dalitz[..].pdf [20 KiB]
HiDef png [181 KiB]
Thumbnail [127 KiB]
*.C file
|

|
Dalitz[..].pdf [23 KiB]
HiDef png [176 KiB]
Thumbnail [125 KiB]
*.C file
|

|
Dalitz[..].pdf [23 KiB]
HiDef png [176 KiB]
Thumbnail [125 KiB]
*.C file
|

|
|
Animated gif made out of all figures.
|
PAPER-2017-010.gif
Thumbnail
|

|